| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
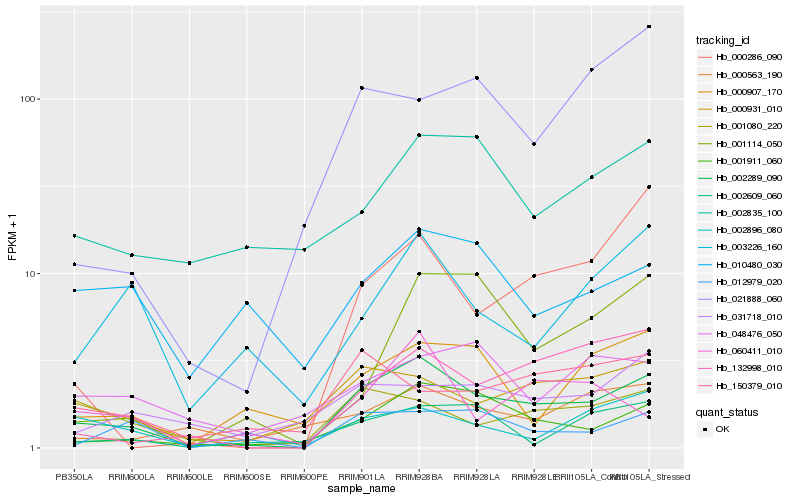
Hb_002289_090 |
0.0 |
- |
- |
cytoplasmic tRNA 2-thiolation protein 2-like [Scleropages formosus] |
| 2 |
Hb_001911_060 |
0.1935540485 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_000931_010 |
0.2180912517 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001080_220 |
0.2366064274 |
- |
- |
TPA: hypothetical protein ZEAMMB73_966926 [Zea mays] |
| 5 |
Hb_012979_020 |
0.236981235 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Solanum lycopersicum] |
| 6 |
Hb_132998_010 |
0.2376771585 |
- |
- |
hypothetical protein JCGZ_00197 [Jatropha curcas] |
| 7 |
Hb_000907_170 |
0.2461959444 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 8 |
Hb_003226_160 |
0.2474392565 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 2-like isoform X2 [Populus euphratica] |
| 9 |
Hb_031718_010 |
0.2516029133 |
- |
- |
- |
| 10 |
Hb_002609_060 |
0.2537235893 |
- |
- |
PREDICTED: sugar transport protein 8-like [Jatropha curcas] |
| 11 |
Hb_150379_010 |
0.2543959464 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 12 |
Hb_002835_100 |
0.2557716257 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |
| 13 |
Hb_000563_190 |
0.2596320645 |
- |
- |
hypothetical protein PRUPE_ppa002828mg [Prunus persica] |
| 14 |
Hb_002896_080 |
0.2596479434 |
- |
- |
- |
| 15 |
Hb_000286_090 |
0.2600198486 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_00197 [Jatropha curcas] |
| 16 |
Hb_060411_010 |
0.2614361799 |
- |
- |
PREDICTED: uncharacterized protein LOC100246093 [Vitis vinifera] |
| 17 |
Hb_048476_050 |
0.2620113723 |
- |
- |
semialdehyde dehydrogenase family protein [Populus trichocarpa] |
| 18 |
Hb_021888_060 |
0.2638721879 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 15 homolog [Jatropha curcas] |
| 19 |
Hb_001114_050 |
0.2645604954 |
- |
- |
hypothetical protein RCOM_0744580 [Ricinus communis] |
| 20 |
Hb_010480_030 |
0.2663659698 |
- |
- |
PREDICTED: LETM1 and EF-hand domain-containing protein 1, mitochondrial-like [Jatropha curcas] |