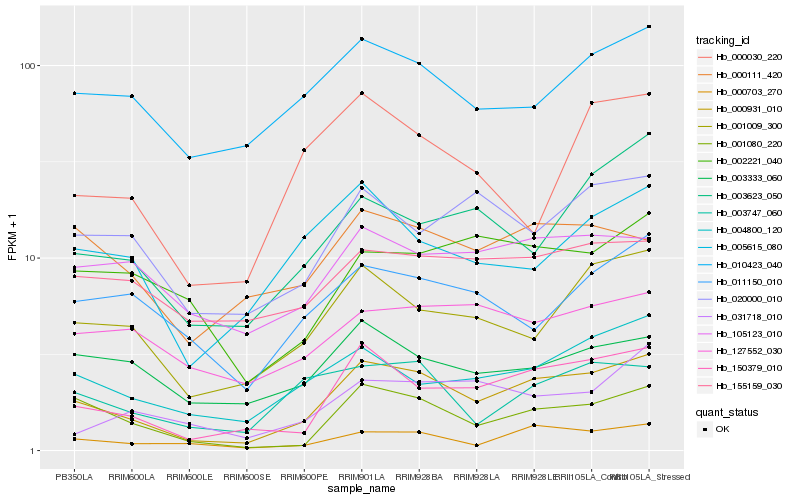
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000931_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_150379_010 |
0.1112035163 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 3 |
Hb_001080_220 |
0.1346147452 |
- |
- |
TPA: hypothetical protein ZEAMMB73_966926 [Zea mays] |
| 4 |
Hb_000703_270 |
0.1436832714 |
- |
- |
hypothetical protein B456_012G039600 [Gossypium raimondii] |
| 5 |
Hb_001009_300 |
0.1479029419 |
transcription factor |
TF Family: GNAT |
PREDICTED: elongator complex protein 3-like [Gossypium raimondii] |
| 6 |
Hb_004800_120 |
0.1524560604 |
- |
- |
PREDICTED: maf-like protein DDB_G0281937 isoform X1 [Jatropha curcas] |
| 7 |
Hb_003623_050 |
0.1674637249 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_020000_010 |
0.1702000684 |
- |
- |
PREDICTED: KH domain-containing protein At5g56140 [Vitis vinifera] |
| 9 |
Hb_005615_080 |
0.1737560644 |
- |
- |
PREDICTED: uncharacterized protein LOC105649609 [Jatropha curcas] |
| 10 |
Hb_003747_060 |
0.1737577619 |
- |
- |
PREDICTED: CMP-N-acetylneuraminate-beta-galactosamide-alpha-2,3-sialyltransferase 2 isoform X2 [Cucumis sativus] |
| 11 |
Hb_010423_040 |
0.174575953 |
- |
- |
hypothetical protein JCGZ_17463 [Jatropha curcas] |
| 12 |
Hb_003333_060 |
0.1783455048 |
- |
- |
PREDICTED: uncharacterized protein LOC105628461 [Jatropha curcas] |
| 13 |
Hb_105123_010 |
0.1788051749 |
- |
- |
PREDICTED: probable protein phosphatase 2C 8 [Jatropha curcas] |
| 14 |
Hb_000111_420 |
0.1798269732 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g80270, mitochondrial-like [Populus euphratica] |
| 15 |
Hb_031718_010 |
0.1822967406 |
- |
- |
- |
| 16 |
Hb_002221_040 |
0.1826510442 |
- |
- |
PREDICTED: uncharacterized protein LOC105639851 [Jatropha curcas] |
| 17 |
Hb_011150_010 |
0.1829797357 |
- |
- |
PREDICTED: uncharacterized RNA-binding protein C1827.05c [Jatropha curcas] |
| 18 |
Hb_127552_030 |
0.1847739118 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 19 |
Hb_000030_220 |
0.1852511026 |
- |
- |
PREDICTED: uncharacterized protein LOC105650405 [Jatropha curcas] |
| 20 |
Hb_155159_030 |
0.1857272085 |
- |
- |
PREDICTED: putative serine/threonine-protein kinase isoform X1 [Jatropha curcas] |