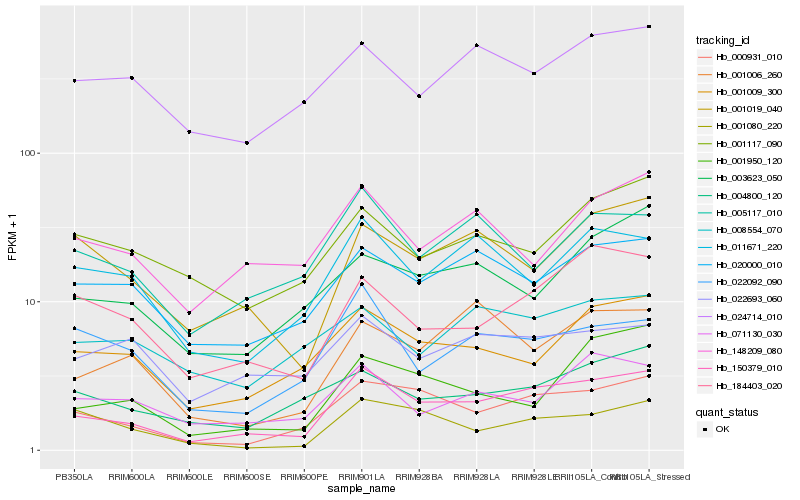
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_150379_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 2 |
Hb_000931_010 |
0.1112035163 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001009_300 |
0.1531758702 |
transcription factor |
TF Family: GNAT |
PREDICTED: elongator complex protein 3-like [Gossypium raimondii] |
| 4 |
Hb_184403_020 |
0.157286178 |
- |
- |
PREDICTED: pre-rRNA-processing protein ESF2 [Vitis vinifera] |
| 5 |
Hb_020000_010 |
0.1574288294 |
- |
- |
PREDICTED: KH domain-containing protein At5g56140 [Vitis vinifera] |
| 6 |
Hb_004800_120 |
0.1624338465 |
- |
- |
PREDICTED: maf-like protein DDB_G0281937 isoform X1 [Jatropha curcas] |
| 7 |
Hb_001019_040 |
0.1660632978 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 8 |
Hb_011671_220 |
0.1661631715 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005117_010 |
0.1695077094 |
- |
- |
PREDICTED: hydroxymethylglutaryl-CoA lyase, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_071130_030 |
0.1699620789 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 17 [Jatropha curcas] |
| 11 |
Hb_022092_090 |
0.1706246223 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 12 |
Hb_003623_050 |
0.1736332892 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001080_220 |
0.1743893645 |
- |
- |
TPA: hypothetical protein ZEAMMB73_966926 [Zea mays] |
| 14 |
Hb_148209_080 |
0.1758716325 |
- |
- |
MED32, putative [Theobroma cacao] |
| 15 |
Hb_001950_120 |
0.1762472242 |
- |
- |
PREDICTED: probable cadmium/zinc-transporting ATPase HMA1, chloroplastic [Jatropha curcas] |
| 16 |
Hb_024714_010 |
0.1769356494 |
- |
- |
HSP20-like chaperones superfamily protein isoform 1 [Theobroma cacao] |
| 17 |
Hb_001006_260 |
0.1782936808 |
- |
- |
PREDICTED: eukaryotic translation initiation factor 3 subunit I-like [Jatropha curcas] |
| 18 |
Hb_022693_060 |
0.179039499 |
- |
- |
- |
| 19 |
Hb_001117_090 |
0.1791046023 |
- |
- |
PREDICTED: NADPH-dependent pterin aldehyde reductase [Jatropha curcas] |
| 20 |
Hb_008554_070 |
0.1818319001 |
- |
- |
PREDICTED: peroxisome biogenesis protein 3-2 isoform X2 [Jatropha curcas] |