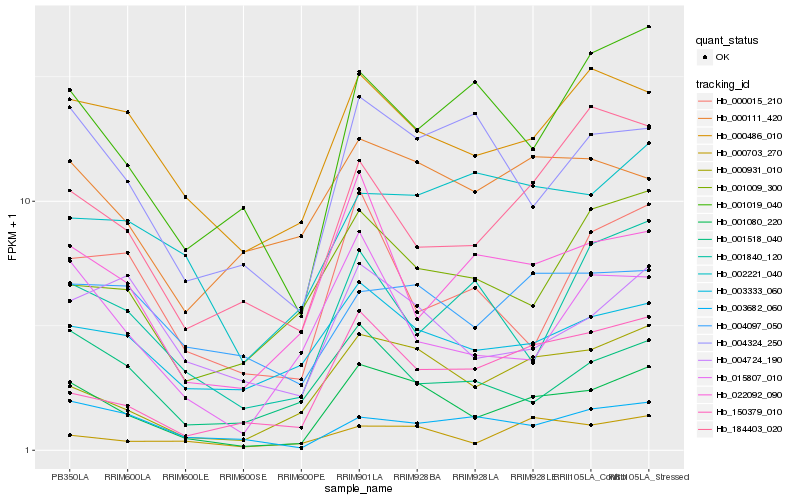
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001080_220 |
0.0 |
- |
- |
TPA: hypothetical protein ZEAMMB73_966926 [Zea mays] |
| 2 |
Hb_000931_010 |
0.1346147452 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_150379_010 |
0.1743893645 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 4 |
Hb_000486_010 |
0.1787093365 |
- |
- |
PREDICTED: uncharacterized protein LOC105646661 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004324_250 |
0.1811650942 |
- |
- |
PREDICTED: protein IWS1 homolog A isoform X1 [Jatropha curcas] |
| 6 |
Hb_000703_270 |
0.1821412775 |
- |
- |
hypothetical protein B456_012G039600 [Gossypium raimondii] |
| 7 |
Hb_003333_060 |
0.1849275276 |
- |
- |
PREDICTED: uncharacterized protein LOC105628461 [Jatropha curcas] |
| 8 |
Hb_001019_040 |
0.186221531 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 9 |
Hb_015807_010 |
0.1873916201 |
- |
- |
hypothetical protein POPTR_0001s44430g [Populus trichocarpa] |
| 10 |
Hb_001518_040 |
0.1899028325 |
- |
- |
PREDICTED: G patch domain-containing protein 11 [Populus euphratica] |
| 11 |
Hb_022092_090 |
0.1919232682 |
- |
- |
PREDICTED: uncharacterized protein LOC105646975 [Jatropha curcas] |
| 12 |
Hb_004724_190 |
0.1931766684 |
- |
- |
NITRILASE-LIKE protein 1 [Populus trichocarpa] |
| 13 |
Hb_001009_300 |
0.1940476466 |
transcription factor |
TF Family: GNAT |
PREDICTED: elongator complex protein 3-like [Gossypium raimondii] |
| 14 |
Hb_003682_060 |
0.1973598844 |
- |
- |
PREDICTED: uncharacterized protein LOC105633833 isoform X1 [Jatropha curcas] |
| 15 |
Hb_184403_020 |
0.1983429239 |
- |
- |
PREDICTED: pre-rRNA-processing protein ESF2 [Vitis vinifera] |
| 16 |
Hb_002221_040 |
0.2011556536 |
- |
- |
PREDICTED: uncharacterized protein LOC105639851 [Jatropha curcas] |
| 17 |
Hb_004097_050 |
0.2012954069 |
- |
- |
PREDICTED: squalene monooxygenase-like [Jatropha curcas] |
| 18 |
Hb_000111_420 |
0.2019866007 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g80270, mitochondrial-like [Populus euphratica] |
| 19 |
Hb_000015_210 |
0.2031969607 |
- |
- |
PREDICTED: uncharacterized protein LOC105647781 [Jatropha curcas] |
| 20 |
Hb_001840_120 |
0.2041886663 |
- |
- |
PREDICTED: uncharacterized protein LOC104228746 [Nicotiana sylvestris] |