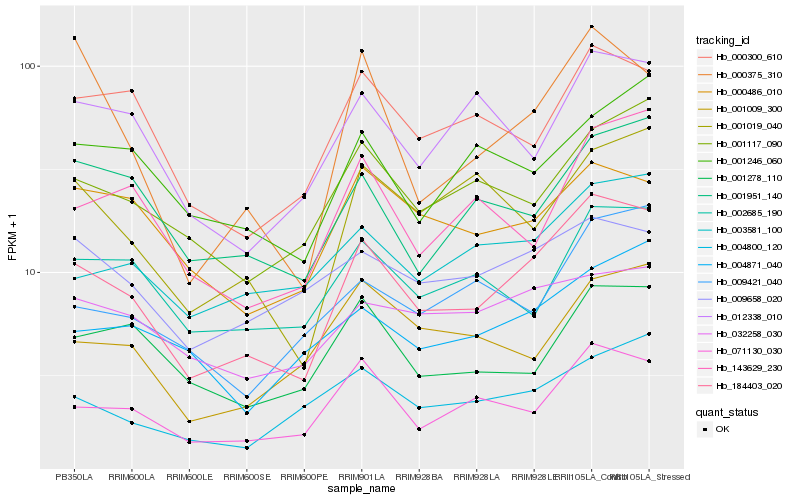
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_184403_020 |
0.0 |
- |
- |
PREDICTED: pre-rRNA-processing protein ESF2 [Vitis vinifera] |
| 2 |
Hb_071130_030 |
0.1164099378 |
- |
- |
PREDICTED: dnaJ homolog subfamily C member 17 [Jatropha curcas] |
| 3 |
Hb_001117_090 |
0.1356497769 |
- |
- |
PREDICTED: NADPH-dependent pterin aldehyde reductase [Jatropha curcas] |
| 4 |
Hb_003581_100 |
0.1369777948 |
- |
- |
PREDICTED: protein TWIN LOV 1 isoform X1 [Jatropha curcas] |
| 5 |
Hb_000300_610 |
0.1374546351 |
- |
- |
cytosolic copper/zinc superoxide dismutase [Hevea brasiliensis] |
| 6 |
Hb_001951_140 |
0.140949477 |
- |
- |
PREDICTED: uncharacterized protein LOC105649757 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002685_190 |
0.1415208209 |
transcription factor |
TF Family: SET |
PREDICTED: probable zinc metalloprotease EGY1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_004800_120 |
0.1415974681 |
- |
- |
PREDICTED: maf-like protein DDB_G0281937 isoform X1 [Jatropha curcas] |
| 9 |
Hb_001278_110 |
0.1417921231 |
- |
- |
PREDICTED: uncharacterized protein LOC104613296 isoform X3 [Nelumbo nucifera] |
| 10 |
Hb_143629_230 |
0.1432545673 |
- |
- |
ring finger, putative [Ricinus communis] |
| 11 |
Hb_001019_040 |
0.1455758219 |
- |
- |
PREDICTED: monothiol glutaredoxin-S7, chloroplastic [Jatropha curcas] |
| 12 |
Hb_012338_010 |
0.1469048416 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_004871_040 |
0.1489675008 |
- |
- |
PREDICTED: tRNA (guanine-N(7)-)-methyltransferase [Jatropha curcas] |
| 14 |
Hb_001009_300 |
0.1491890544 |
transcription factor |
TF Family: GNAT |
PREDICTED: elongator complex protein 3-like [Gossypium raimondii] |
| 15 |
Hb_009421_040 |
0.1497197353 |
- |
- |
PREDICTED: eukaryotic initiation factor 4A [Jatropha curcas] |
| 16 |
Hb_000486_010 |
0.1502189435 |
- |
- |
PREDICTED: uncharacterized protein LOC105646661 isoform X1 [Jatropha curcas] |
| 17 |
Hb_009658_020 |
0.1511819328 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 15 [Jatropha curcas] |
| 18 |
Hb_032258_030 |
0.151318748 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC105647563 [Jatropha curcas] |
| 19 |
Hb_001246_060 |
0.1514564687 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 20 |
Hb_000375_310 |
0.1520906134 |
- |
- |
hypothetical protein JCGZ_18643 [Jatropha curcas] |