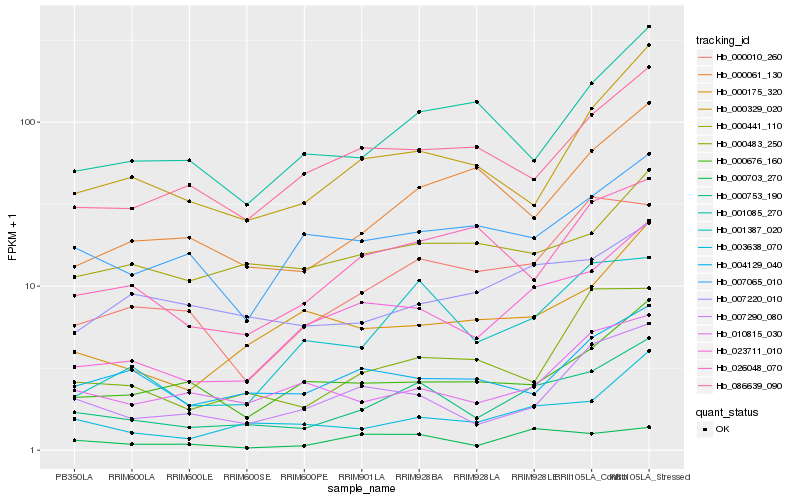
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000753_190 |
0.0 |
- |
- |
PREDICTED: D-aminoacyl-tRNA deacylase [Populus euphratica] |
| 2 |
Hb_023711_010 |
0.1259690568 |
- |
- |
- |
| 3 |
Hb_003638_070 |
0.134683244 |
- |
- |
hypothetical protein EUGRSUZ_E00638 [Eucalyptus grandis] |
| 4 |
Hb_007290_080 |
0.1572616984 |
- |
- |
hypothetical protein CISIN_1g047098mg [Citrus sinensis] |
| 5 |
Hb_000010_260 |
0.1580878929 |
- |
- |
PREDICTED: uncharacterized protein LOC105640273 [Jatropha curcas] |
| 6 |
Hb_000329_020 |
0.1604153496 |
- |
- |
cytoplasmic ribosomal protein S13 [Panax ginseng] |
| 7 |
Hb_001387_020 |
0.1623170494 |
- |
- |
PREDICTED: uncharacterized protein LOC102587892 [Solanum tuberosum] |
| 8 |
Hb_086639_090 |
0.1643731844 |
- |
- |
60S ribosomal protein L7a, putative [Ricinus communis] |
| 9 |
Hb_010815_030 |
0.1659455347 |
- |
- |
PREDICTED: (-)-germacrene D synthase [Vitis vinifera] |
| 10 |
Hb_000061_130 |
0.1660275146 |
- |
- |
rotamase, putative [Ricinus communis] |
| 11 |
Hb_000483_250 |
0.1673778969 |
- |
- |
PREDICTED: uncharacterized protein LOC105803540 [Gossypium raimondii] |
| 12 |
Hb_000676_160 |
0.167888154 |
- |
- |
folate carrier protein, putative [Ricinus communis] |
| 13 |
Hb_000175_320 |
0.1694145392 |
- |
- |
PREDICTED: formin-like protein 14 [Jatropha curcas] |
| 14 |
Hb_007220_010 |
0.1702693877 |
- |
- |
PREDICTED: methylcrotonoyl-CoA carboxylase beta chain, mitochondrial [Jatropha curcas] |
| 15 |
Hb_026048_070 |
0.1703445404 |
- |
- |
PREDICTED: uncharacterized protein LOC105645736 [Jatropha curcas] |
| 16 |
Hb_000441_110 |
0.1705377955 |
- |
- |
PREDICTED: uncharacterized protein LOC105646523 [Jatropha curcas] |
| 17 |
Hb_000703_270 |
0.1707233897 |
- |
- |
hypothetical protein B456_012G039600 [Gossypium raimondii] |
| 18 |
Hb_001085_270 |
0.1709871385 |
- |
- |
60S ribosomal protein L13aA [Hevea brasiliensis] |
| 19 |
Hb_007065_010 |
0.1714767923 |
- |
- |
hypothetical protein Csa_3G379730 [Cucumis sativus] |
| 20 |
Hb_004129_040 |
0.1720582659 |
- |
- |
PREDICTED: protein VACUOLELESS1 [Vitis vinifera] |