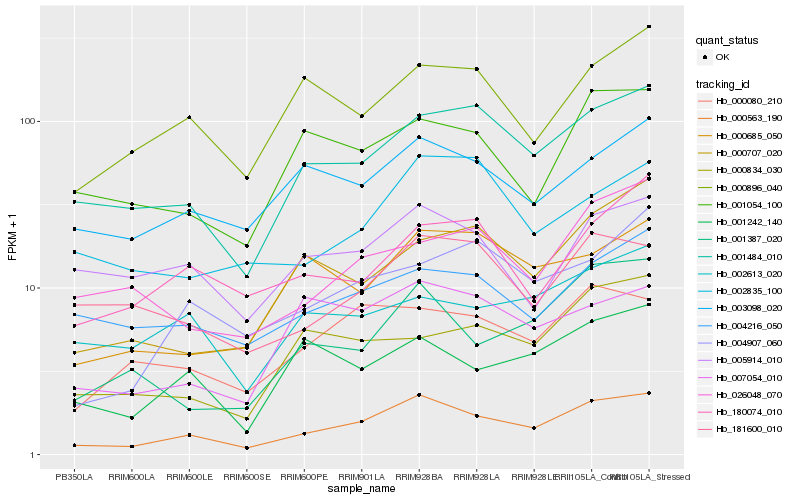
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000563_190 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa002828mg [Prunus persica] |
| 2 |
Hb_001484_010 |
0.1218382585 |
- |
- |
hypothetical protein POPTR_0005s07440g [Populus trichocarpa] |
| 3 |
Hb_000080_210 |
0.1446343423 |
- |
- |
PREDICTED: uncharacterized protein LOC105628422 [Jatropha curcas] |
| 4 |
Hb_003098_020 |
0.1507203311 |
- |
- |
40S ribosomal protein S5B [Hevea brasiliensis] |
| 5 |
Hb_001387_020 |
0.1528385331 |
- |
- |
PREDICTED: uncharacterized protein LOC102587892 [Solanum tuberosum] |
| 6 |
Hb_000896_040 |
0.1540327671 |
- |
- |
BnaC02g13240D [Brassica napus] |
| 7 |
Hb_007054_010 |
0.1559036897 |
- |
- |
PREDICTED: WD repeat-containing protein 82 [Jatropha curcas] |
| 8 |
Hb_001054_100 |
0.1560679624 |
- |
- |
PREDICTED: uncharacterized protein LOC105634642 [Jatropha curcas] |
| 9 |
Hb_001242_140 |
0.1566542548 |
- |
- |
mitochondrial carrier protein, putative [Ricinus communis] |
| 10 |
Hb_004907_060 |
0.1574153812 |
- |
- |
PREDICTED: tropinone reductase homolog At5g06060-like [Jatropha curcas] |
| 11 |
Hb_004216_050 |
0.1578967017 |
- |
- |
PREDICTED: uncharacterized protein LOC105628422 [Jatropha curcas] |
| 12 |
Hb_000685_050 |
0.1591762201 |
- |
- |
PREDICTED: mitochondrial inner membrane protein OXA1-like [Populus euphratica] |
| 13 |
Hb_000707_020 |
0.1594564111 |
- |
- |
hypothetical protein B456_002G255500 [Gossypium raimondii] |
| 14 |
Hb_181600_010 |
0.1594941702 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 27 [Jatropha curcas] |
| 15 |
Hb_180074_010 |
0.1643455656 |
- |
- |
Single-stranded DNA-binding protein, mitochondrial precursor, putative [Ricinus communis] |
| 16 |
Hb_005914_010 |
0.164673801 |
- |
- |
PREDICTED: peroxisomal membrane protein PMP22 [Jatropha curcas] |
| 17 |
Hb_000834_030 |
0.1652023692 |
- |
- |
PREDICTED: protein Mpv17 isoform X2 [Jatropha curcas] |
| 18 |
Hb_026048_070 |
0.1652059245 |
- |
- |
PREDICTED: uncharacterized protein LOC105645736 [Jatropha curcas] |
| 19 |
Hb_002835_100 |
0.1654250496 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |
| 20 |
Hb_002613_020 |
0.1673346306 |
- |
- |
ADP-ribose pyrophosphatase, putative [Ricinus communis] |