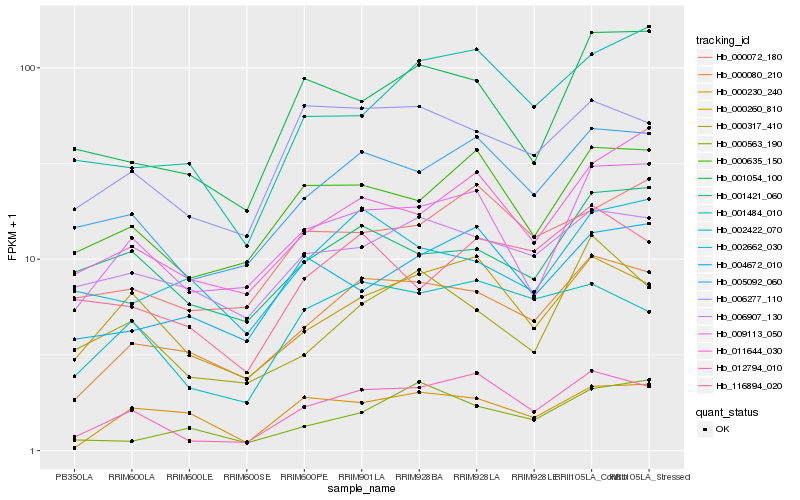
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000080_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105628422 [Jatropha curcas] |
| 2 |
Hb_005092_060 |
0.1119627743 |
- |
- |
snf1-kinase beta subunit, plants, putative [Ricinus communis] |
| 3 |
Hb_009113_050 |
0.118611273 |
- |
- |
PREDICTED: uncharacterized protein LOC105644587 [Jatropha curcas] |
| 4 |
Hb_006907_130 |
0.121084263 |
- |
- |
unnamed protein product [Coffea canephora] |
| 5 |
Hb_012794_010 |
0.1238126132 |
- |
- |
PREDICTED: probable RNA helicase SDE3 [Populus euphratica] |
| 6 |
Hb_006277_110 |
0.1305804013 |
- |
- |
protein with unknown function [Ricinus communis] |
| 7 |
Hb_002662_030 |
0.1319533369 |
- |
- |
PREDICTED: mannose-P-dolichol utilization defect 1 protein homolog 2 [Jatropha curcas] |
| 8 |
Hb_000260_810 |
0.1337017754 |
- |
- |
- |
| 9 |
Hb_000635_150 |
0.137848469 |
- |
- |
mitochondrial import receptor subunit TOM20-2 family protein [Populus trichocarpa] |
| 10 |
Hb_000230_240 |
0.1384085499 |
- |
- |
tRNA/rRNA methyltransferase family protein [Populus trichocarpa] |
| 11 |
Hb_011644_030 |
0.1423393867 |
- |
- |
PREDICTED: 50S ribosomal protein L12, cyanelle-like [Pyrus x bretschneideri] |
| 12 |
Hb_002422_070 |
0.1436081704 |
- |
- |
PREDICTED: putative protein TPRXL [Malus domestica] |
| 13 |
Hb_000563_190 |
0.1446343423 |
- |
- |
hypothetical protein PRUPE_ppa002828mg [Prunus persica] |
| 14 |
Hb_000072_180 |
0.1452164243 |
- |
- |
PREDICTED: EVI5-like protein [Jatropha curcas] |
| 15 |
Hb_116894_020 |
0.146058214 |
- |
- |
hypothetical protein POPTR_0008s07660g [Populus trichocarpa] |
| 16 |
Hb_004672_010 |
0.1463571682 |
- |
- |
ecotropic viral integration site, putative [Ricinus communis] |
| 17 |
Hb_000317_410 |
0.1464637884 |
- |
- |
PREDICTED: UDP-glycosyltransferase 73C5-like [Jatropha curcas] |
| 18 |
Hb_001054_100 |
0.1475759374 |
- |
- |
PREDICTED: uncharacterized protein LOC105634642 [Jatropha curcas] |
| 19 |
Hb_001484_010 |
0.1488647671 |
- |
- |
hypothetical protein POPTR_0005s07440g [Populus trichocarpa] |
| 20 |
Hb_001421_060 |
0.1495272757 |
- |
- |
PREDICTED: tobamovirus multiplication protein 2B isoform X2 [Jatropha curcas] |