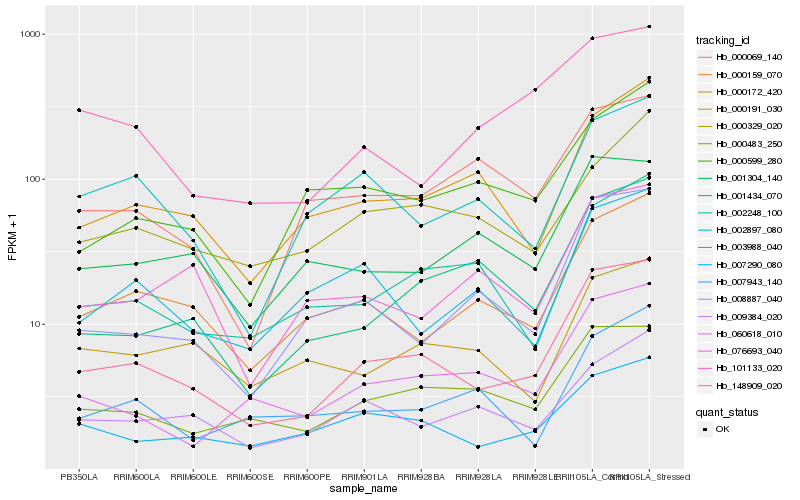
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_148909_020 |
0.0 |
- |
- |
PREDICTED: glutamyl-tRNA(Gln) amidotransferase subunit C, chloroplastic/mitochondrial [Jatropha curcas] |
| 2 |
Hb_000483_250 |
0.1349485339 |
- |
- |
PREDICTED: uncharacterized protein LOC105803540 [Gossypium raimondii] |
| 3 |
Hb_000159_070 |
0.1375705571 |
- |
- |
JHL03K20.4 [Jatropha curcas] |
| 4 |
Hb_007290_080 |
0.1392548654 |
- |
- |
hypothetical protein CISIN_1g047098mg [Citrus sinensis] |
| 5 |
Hb_008887_040 |
0.1401395608 |
- |
- |
Ribonuclease P family protein / Rpp14 family protein isoform 1 [Theobroma cacao] |
| 6 |
Hb_060618_010 |
0.1470725223 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 7 |
Hb_000172_420 |
0.1490013843 |
- |
- |
60S ribosomal protein L30, putative [Ricinus communis] |
| 8 |
Hb_001434_070 |
0.1503337064 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM8 [Jatropha curcas] |
| 9 |
Hb_002248_100 |
0.1530176894 |
- |
- |
hypothetical protein VITISV_027197 [Vitis vinifera] |
| 10 |
Hb_009384_020 |
0.1537687765 |
- |
- |
hypothetical protein JCGZ_24168 [Jatropha curcas] |
| 11 |
Hb_001304_140 |
0.1571372355 |
- |
- |
PREDICTED: transmembrane protein 256 homolog [Jatropha curcas] |
| 12 |
Hb_002897_080 |
0.1579388582 |
- |
- |
hypothetical protein JCGZ_03306 [Jatropha curcas] |
| 13 |
Hb_076693_040 |
0.1639088231 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B [Tarenaya hassleriana] |
| 14 |
Hb_000191_030 |
0.1655451862 |
- |
- |
hypothetical protein POPTR_0012s07830g [Populus trichocarpa] |
| 15 |
Hb_000599_280 |
0.1673749319 |
- |
- |
PREDICTED: 60S ribosomal protein L14-1 [Jatropha curcas] |
| 16 |
Hb_003988_040 |
0.1687484192 |
- |
- |
PREDICTED: uncharacterized protein LOC105633684 [Jatropha curcas] |
| 17 |
Hb_000329_020 |
0.1693380058 |
- |
- |
cytoplasmic ribosomal protein S13 [Panax ginseng] |
| 18 |
Hb_101133_020 |
0.1697137848 |
- |
- |
PREDICTED: 15.7 kDa heat shock protein, peroxisomal [Jatropha curcas] |
| 19 |
Hb_000069_140 |
0.1713122874 |
- |
- |
PREDICTED: uncharacterized protein LOC105642831 [Jatropha curcas] |
| 20 |
Hb_007943_140 |
0.1726795881 |
- |
- |
PREDICTED: uncharacterized protein LOC103340977 [Prunus mume] |