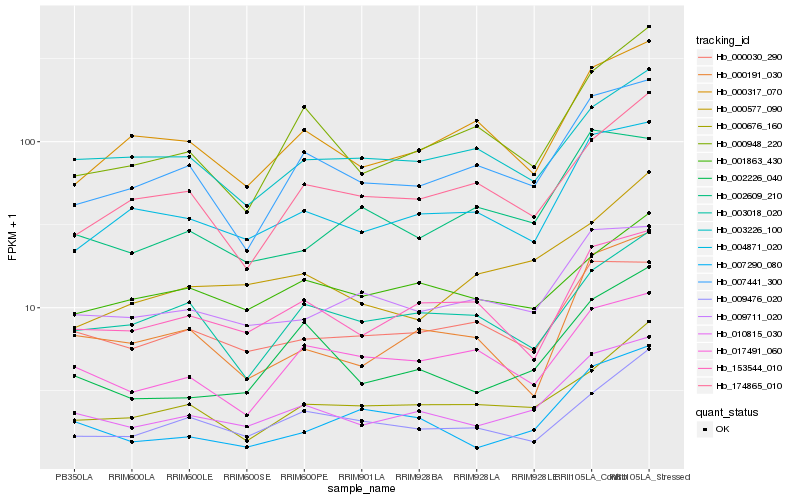
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010815_030 |
0.0 |
- |
- |
PREDICTED: (-)-germacrene D synthase [Vitis vinifera] |
| 2 |
Hb_004871_020 |
0.0970398466 |
transcription factor |
TF Family: SWI/SNF-BAF60b |
PREDICTED: protein TRI1 [Jatropha curcas] |
| 3 |
Hb_007441_300 |
0.1016798476 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 4 |
Hb_002226_040 |
0.1019979194 |
- |
- |
PREDICTED: uncharacterized protein LOC105641318 isoform X2 [Jatropha curcas] |
| 5 |
Hb_009711_020 |
0.1067974605 |
- |
- |
PREDICTED: uncharacterized protein LOC105647794 isoform X2 [Jatropha curcas] |
| 6 |
Hb_153544_010 |
0.1088052621 |
- |
- |
PREDICTED: coiled-coil domain-containing protein 130-like [Populus euphratica] |
| 7 |
Hb_000030_290 |
0.1089778807 |
- |
- |
PREDICTED: uncharacterized protein LOC100247068 [Vitis vinifera] |
| 8 |
Hb_000676_160 |
0.1137422236 |
- |
- |
folate carrier protein, putative [Ricinus communis] |
| 9 |
Hb_007290_080 |
0.1187968427 |
- |
- |
hypothetical protein CISIN_1g047098mg [Citrus sinensis] |
| 10 |
Hb_000948_220 |
0.1195527431 |
- |
- |
hypothetical protein CICLE_v10013189mg [Citrus clementina] |
| 11 |
Hb_000317_070 |
0.119688586 |
- |
- |
PREDICTED: uncharacterized protein LOC103942205 isoform X2 [Pyrus x bretschneideri] |
| 12 |
Hb_003226_100 |
0.121965561 |
- |
- |
PREDICTED: 60S ribosomal protein L18-2 [Jatropha curcas] |
| 13 |
Hb_009476_020 |
0.1230787356 |
- |
- |
tryptophan biosynthesis protein, putative [Ricinus communis] |
| 14 |
Hb_000577_090 |
0.1245884537 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_000191_030 |
0.1251798135 |
- |
- |
hypothetical protein POPTR_0012s07830g [Populus trichocarpa] |
| 16 |
Hb_002609_210 |
0.1256220206 |
- |
- |
PREDICTED: pleckstrin homology domain-containing protein 1 [Jatropha curcas] |
| 17 |
Hb_001863_430 |
0.128162208 |
- |
- |
PREDICTED: probable protein phosphatase 2C 9 [Populus euphratica] |
| 18 |
Hb_003018_020 |
0.128175305 |
- |
- |
PREDICTED: uncharacterized protein LOC105628697 [Jatropha curcas] |
| 19 |
Hb_017491_060 |
0.1285349193 |
- |
- |
PREDICTED: probable complex I intermediate-associated protein 30 isoform X2 [Jatropha curcas] |
| 20 |
Hb_174865_010 |
0.1288287583 |
- |
- |
transcription initiation factor iia (tfiia), gamma chain, putative [Ricinus communis] |