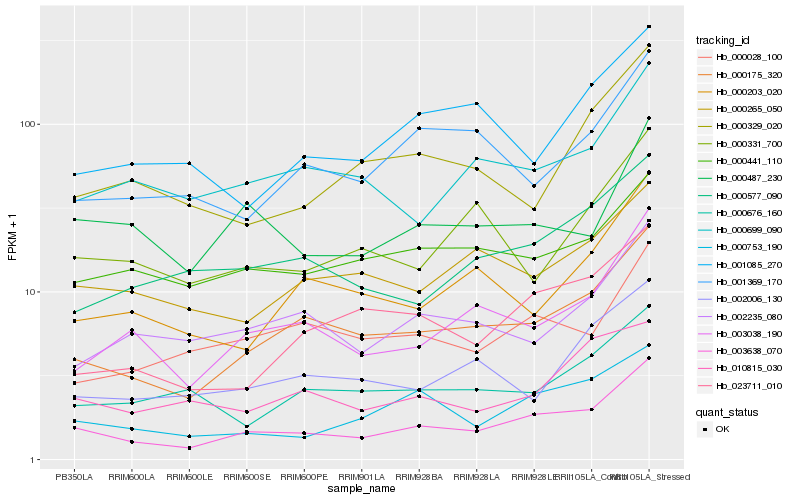
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003638_070 |
0.0 |
- |
- |
hypothetical protein EUGRSUZ_E00638 [Eucalyptus grandis] |
| 2 |
Hb_000175_320 |
0.0988190579 |
- |
- |
PREDICTED: formin-like protein 14 [Jatropha curcas] |
| 3 |
Hb_003038_190 |
0.1345603966 |
transcription factor |
TF Family: C2H2 |
PREDICTED: uncharacterized protein LOC105645961 [Jatropha curcas] |
| 4 |
Hb_000753_190 |
0.134683244 |
- |
- |
PREDICTED: D-aminoacyl-tRNA deacylase [Populus euphratica] |
| 5 |
Hb_023711_010 |
0.1475473467 |
- |
- |
- |
| 6 |
Hb_000577_090 |
0.1487848835 |
- |
- |
PREDICTED: 50S ribosomal protein L12-1, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_000487_230 |
0.1493636731 |
- |
- |
PREDICTED: probable BOI-related E3 ubiquitin-protein ligase 2 [Jatropha curcas] |
| 8 |
Hb_000699_090 |
0.1516577082 |
- |
- |
cyclophilin, putative [Ricinus communis] |
| 9 |
Hb_002235_080 |
0.1537175599 |
- |
- |
PREDICTED: U3 small nucleolar ribonucleoprotein protein IMP3 [Vitis vinifera] |
| 10 |
Hb_000028_100 |
0.1548480568 |
transcription factor |
TF Family: M-type |
PREDICTED: agamous-like MADS-box protein AGL61 [Jatropha curcas] |
| 11 |
Hb_000265_050 |
0.1561109503 |
- |
- |
hypothetical protein CISIN_1g033530mg [Citrus sinensis] |
| 12 |
Hb_000441_110 |
0.1574018797 |
- |
- |
PREDICTED: uncharacterized protein LOC105646523 [Jatropha curcas] |
| 13 |
Hb_000203_020 |
0.1598593386 |
- |
- |
PREDICTED: 2-aminoethanethiol dioxygenase-like [Jatropha curcas] |
| 14 |
Hb_000676_160 |
0.1610178932 |
- |
- |
folate carrier protein, putative [Ricinus communis] |
| 15 |
Hb_002006_130 |
0.1618028505 |
- |
- |
PREDICTED: uncharacterized protein LOC105648712 [Jatropha curcas] |
| 16 |
Hb_010815_030 |
0.1624273049 |
- |
- |
PREDICTED: (-)-germacrene D synthase [Vitis vinifera] |
| 17 |
Hb_000329_020 |
0.1624857486 |
- |
- |
cytoplasmic ribosomal protein S13 [Panax ginseng] |
| 18 |
Hb_001369_170 |
0.1629193064 |
- |
- |
PREDICTED: 60S ribosomal protein L15-1-like [Gossypium raimondii] |
| 19 |
Hb_000331_700 |
0.1674243154 |
- |
- |
PREDICTED: uncharacterized protein LOC105640277 [Jatropha curcas] |
| 20 |
Hb_001085_270 |
0.170060474 |
- |
- |
60S ribosomal protein L13aA [Hevea brasiliensis] |