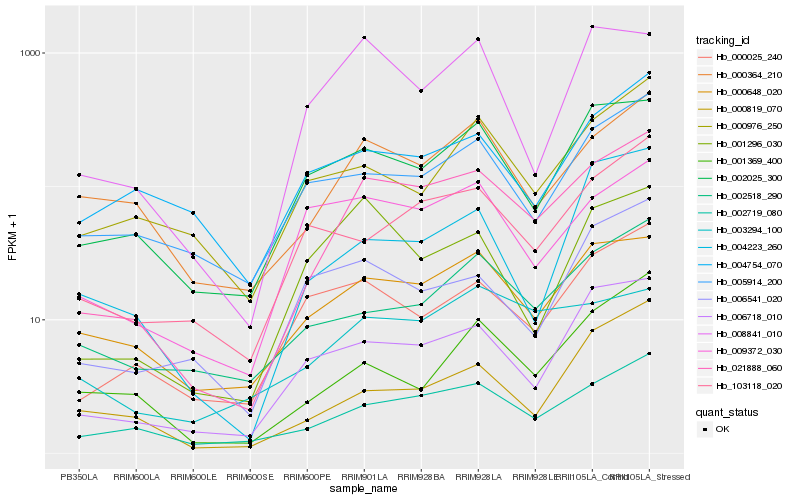
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_021888_060 |
0.0 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 15 homolog [Jatropha curcas] |
| 2 |
Hb_002719_080 |
0.1393933906 |
- |
- |
hypothetical protein POPTR_0006s27920g [Populus trichocarpa] |
| 3 |
Hb_006718_010 |
0.1590378227 |
- |
- |
PREDICTED: histone H2AX-like [Elaeis guineensis] |
| 4 |
Hb_000648_020 |
0.1632080649 |
- |
- |
PREDICTED: chaperone protein dnaJ 10-like [Jatropha curcas] |
| 5 |
Hb_000364_210 |
0.1649948739 |
- |
- |
Mitochondrial import receptor subunit TOM7-1, putative [Ricinus communis] |
| 6 |
Hb_002025_300 |
0.1652370732 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] 1 beta subcomplex subunit 7 [Jatropha curcas] |
| 7 |
Hb_103118_020 |
0.1762958634 |
- |
- |
PREDICTED: protein mago nashi homolog [Jatropha curcas] |
| 8 |
Hb_000025_240 |
0.1789706537 |
- |
- |
PREDICTED: uncharacterized protein LOC105628964 [Jatropha curcas] |
| 9 |
Hb_002518_290 |
0.181277039 |
- |
- |
Acyl-coenzyme A thioesterase 13 [Gossypium arboreum] |
| 10 |
Hb_009372_030 |
0.18347772 |
- |
- |
50S ribosomal protein L6, putative [Ricinus communis] |
| 11 |
Hb_003294_100 |
0.1859403268 |
- |
- |
hypothetical protein VITISV_009629 [Vitis vinifera] |
| 12 |
Hb_005914_200 |
0.1870995306 |
- |
- |
PREDICTED: 60S ribosomal protein L23-like isoform X1 [Tarenaya hassleriana] |
| 13 |
Hb_001296_030 |
0.1875203623 |
- |
- |
PREDICTED: uncharacterized protein LOC105638276 [Jatropha curcas] |
| 14 |
Hb_008841_010 |
0.1880812416 |
- |
- |
PREDICTED: uncharacterized protein LOC103323653 [Prunus mume] |
| 15 |
Hb_001369_400 |
0.1933231782 |
- |
- |
PREDICTED: uncharacterized protein LOC105648609 [Jatropha curcas] |
| 16 |
Hb_004223_260 |
0.1959759046 |
- |
- |
hypothetical protein F383_09277 [Gossypium arboreum] |
| 17 |
Hb_000819_070 |
0.1971118723 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 1 [Jatropha curcas] |
| 18 |
Hb_000976_250 |
0.1987692513 |
- |
- |
- |
| 19 |
Hb_006541_020 |
0.199907001 |
- |
- |
unknown [Lotus japonicus] |
| 20 |
Hb_004754_070 |
0.203406257 |
- |
- |
PREDICTED: 60S ribosomal protein L30-like [Gossypium raimondii] |