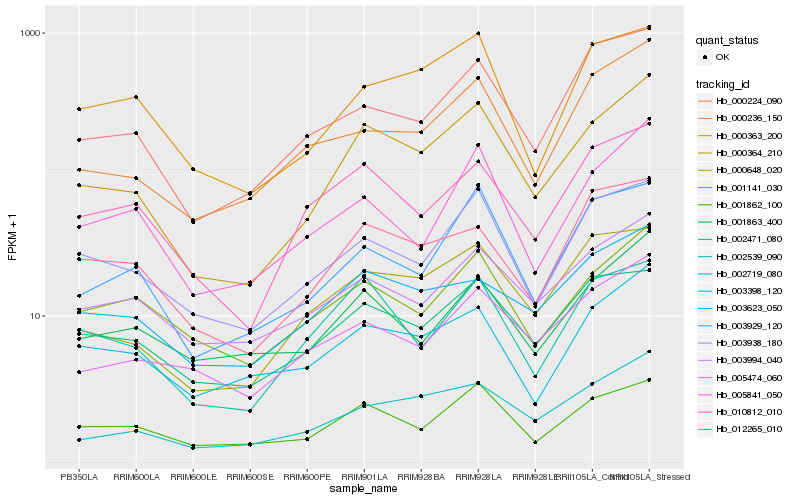
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000364_210 |
0.0 |
- |
- |
Mitochondrial import receptor subunit TOM7-1, putative [Ricinus communis] |
| 2 |
Hb_001862_100 |
0.1086249131 |
- |
- |
PREDICTED: uncharacterized protein LOC105639799 isoform X2 [Jatropha curcas] |
| 3 |
Hb_002719_080 |
0.1096407807 |
- |
- |
hypothetical protein POPTR_0006s27920g [Populus trichocarpa] |
| 4 |
Hb_000648_020 |
0.116806772 |
- |
- |
PREDICTED: chaperone protein dnaJ 10-like [Jatropha curcas] |
| 5 |
Hb_003938_180 |
0.1216008933 |
- |
- |
lesion inducing family protein [Populus trichocarpa] |
| 6 |
Hb_002539_090 |
0.1268985921 |
- |
- |
PREDICTED: uncharacterized protein LOC103489178 [Cucumis melo] |
| 7 |
Hb_003929_120 |
0.1270670691 |
- |
- |
PREDICTED: 50S ribosomal protein L3-2, chloroplastic [Jatropha curcas] |
| 8 |
Hb_010812_010 |
0.1279479787 |
- |
- |
hypothetical protein JCGZ_03306 [Jatropha curcas] |
| 9 |
Hb_000224_090 |
0.1286548933 |
- |
- |
Eukaryotic translation initiation factor 1A [Gossypium arboreum] |
| 10 |
Hb_012265_010 |
0.130235148 |
- |
- |
hypothetical protein CISIN_1g026883mg [Citrus sinensis] |
| 11 |
Hb_003994_040 |
0.1308771651 |
- |
- |
- |
| 12 |
Hb_002471_080 |
0.131072895 |
- |
- |
oxidoreductase, putative [Ricinus communis] |
| 13 |
Hb_003623_050 |
0.1311216786 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000363_200 |
0.1312999029 |
- |
- |
40S ribosomal protein S8, putative [Ricinus communis] |
| 15 |
Hb_005474_060 |
0.1314653084 |
- |
- |
hypothetical protein PHAVU_011G019700g [Phaseolus vulgaris] |
| 16 |
Hb_000236_150 |
0.1315071786 |
- |
- |
glycine-rich RNA-binding protein, putative [Ricinus communis] |
| 17 |
Hb_005841_050 |
0.1343684904 |
- |
- |
PREDICTED: F-box protein SKIP1-like [Jatropha curcas] |
| 18 |
Hb_003398_120 |
0.1344068471 |
- |
- |
PREDICTED: uncharacterized protein LOC105641067 [Jatropha curcas] |
| 19 |
Hb_001863_400 |
0.1346237399 |
- |
- |
hypothetical protein EUGRSUZ_F03302 [Eucalyptus grandis] |
| 20 |
Hb_001141_030 |
0.1346452317 |
- |
- |
hypothetical protein PRUPE_ppa024854mg [Prunus persica] |