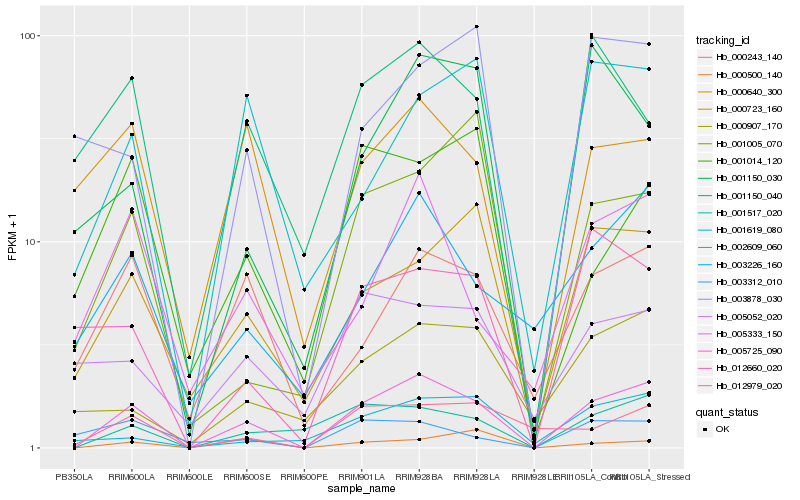
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005725_090 |
0.0 |
- |
- |
PREDICTED: phytosulfokines 3 [Cicer arietinum] |
| 2 |
Hb_001517_020 |
0.2122524221 |
- |
- |
PREDICTED: probable ATP-dependent RNA helicase DDX11 [Jatropha curcas] |
| 3 |
Hb_000243_140 |
0.2409545503 |
transcription factor |
TF Family: SRS |
Lateral root primordium protein-related, putative [Theobroma cacao] |
| 4 |
Hb_000723_160 |
0.2423482561 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP11 [Jatropha curcas] |
| 5 |
Hb_012979_020 |
0.2469113544 |
- |
- |
PREDICTED: neutral alpha-glucosidase C [Solanum lycopersicum] |
| 6 |
Hb_003226_160 |
0.2469722228 |
- |
- |
PREDICTED: poly [ADP-ribose] polymerase 2-like isoform X2 [Populus euphratica] |
| 7 |
Hb_005333_150 |
0.2521487792 |
- |
- |
hypothetical protein POPTR_0002s10520g [Populus trichocarpa] |
| 8 |
Hb_000907_170 |
0.256013942 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_001619_080 |
0.2562551633 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_002609_060 |
0.2592901014 |
- |
- |
PREDICTED: sugar transport protein 8-like [Jatropha curcas] |
| 11 |
Hb_001150_040 |
0.259336693 |
- |
- |
PREDICTED: 2-alkenal reductase (NADP(+)-dependent)-like [Solanum lycopersicum] |
| 12 |
Hb_000640_300 |
0.2609821803 |
transcription factor |
TF Family: WRKY |
WRKY1 [Hevea brasiliensis] |
| 13 |
Hb_012660_020 |
0.2636302738 |
- |
- |
- |
| 14 |
Hb_000500_140 |
0.2654006651 |
transcription factor |
TF Family: MYB-related |
PREDICTED: protein REVEILLE 4 [Jatropha curcas] |
| 15 |
Hb_001150_030 |
0.2685161735 |
- |
- |
NADP-dependent oxidoreductase P1 isoform 3 [Theobroma cacao] |
| 16 |
Hb_003878_030 |
0.2686825332 |
- |
- |
hypothetical protein POPTR_0011s11120g [Populus trichocarpa] |
| 17 |
Hb_005052_020 |
0.2729418271 |
- |
- |
- |
| 18 |
Hb_001005_070 |
0.2732393626 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor ERF113-like [Jatropha curcas] |
| 19 |
Hb_001014_120 |
0.2739848573 |
transcription factor |
TF Family: ERF |
ethylene-responsive transcription factor ERF113 [Jatropha curcas] |
| 20 |
Hb_003312_010 |
0.2758002226 |
- |
- |
PREDICTED: uncharacterized protein LOC104428797 [Eucalyptus grandis] |