| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
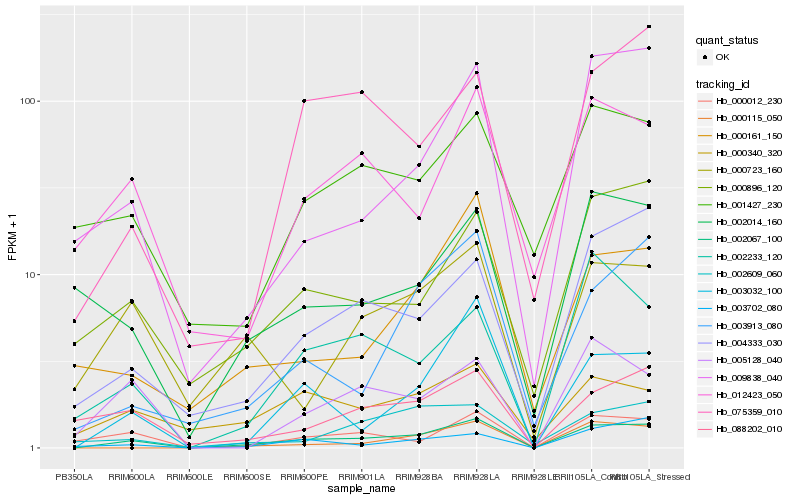
Hb_002067_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_003032_100 |
0.1975463894 |
- |
- |
PREDICTED: GDP-mannose transporter GONST4 [Jatropha curcas] |
| 3 |
Hb_000012_230 |
0.1998356968 |
- |
- |
PREDICTED: uncharacterized protein LOC105629874 [Jatropha curcas] |
| 4 |
Hb_004333_030 |
0.2064840176 |
- |
- |
PREDICTED: vacuolar protein sorting-associated protein 26A [Jatropha curcas] |
| 5 |
Hb_000896_120 |
0.208622303 |
- |
- |
PREDICTED: acyl-protein thioesterase 2-like [Populus euphratica] |
| 6 |
Hb_088202_010 |
0.2095577819 |
- |
- |
hypothetical protein JCGZ_13241 [Jatropha curcas] |
| 7 |
Hb_000115_050 |
0.2097167137 |
- |
- |
- |
| 8 |
Hb_000723_160 |
0.2137883369 |
transcription factor |
TF Family: TCP |
PREDICTED: transcription factor TCP11 [Jatropha curcas] |
| 9 |
Hb_002609_060 |
0.2153749745 |
- |
- |
PREDICTED: sugar transport protein 8-like [Jatropha curcas] |
| 10 |
Hb_002233_120 |
0.2170141537 |
- |
- |
- |
| 11 |
Hb_012423_050 |
0.2177672485 |
- |
- |
PREDICTED: transmembrane protein 64 [Jatropha curcas] |
| 12 |
Hb_003702_080 |
0.2194042457 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 13 |
Hb_005128_040 |
0.2200220699 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000340_320 |
0.220122018 |
- |
- |
PREDICTED: uncharacterized protein LOC105650052 [Jatropha curcas] |
| 15 |
Hb_003913_080 |
0.2213962341 |
- |
- |
rpl27A [Hemiselmis andersenii] |
| 16 |
Hb_009838_040 |
0.2221181174 |
- |
- |
hypothetical protein JCGZ_12866 [Jatropha curcas] |
| 17 |
Hb_002014_160 |
0.2238528826 |
transcription factor |
TF Family: HSF |
PREDICTED: heat stress transcription factor C-1 [Jatropha curcas] |
| 18 |
Hb_075359_010 |
0.2242024132 |
- |
- |
eukaryotic translation initiation factor 2c, putative [Ricinus communis] |
| 19 |
Hb_001427_230 |
0.2242198608 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000161_150 |
0.2243544156 |
- |
- |
PREDICTED: protein argonaute 2-like [Jatropha curcas] |