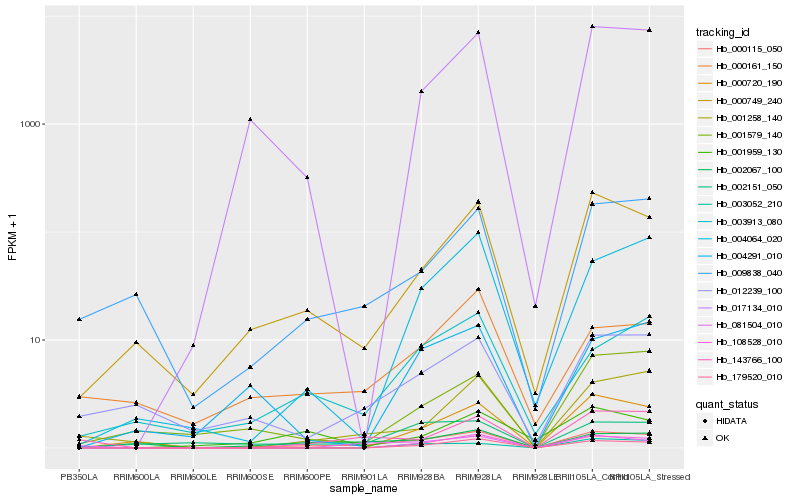
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000115_050 |
0.0 |
- |
- |
- |
| 2 |
Hb_000749_240 |
0.166432463 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_017134_010 |
0.1886673799 |
- |
- |
HEV1.2 [Hevea brasiliensis] |
| 4 |
Hb_003913_080 |
0.2096734277 |
- |
- |
rpl27A [Hemiselmis andersenii] |
| 5 |
Hb_002067_100 |
0.2097167137 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_002151_050 |
0.2131743935 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 7 |
Hb_108528_010 |
0.218385863 |
- |
- |
unnamed protein product [Coffea canephora] |
| 8 |
Hb_009838_040 |
0.2199659311 |
- |
- |
hypothetical protein JCGZ_12866 [Jatropha curcas] |
| 9 |
Hb_012239_100 |
0.2203725232 |
- |
- |
hypothetical protein VITISV_043897 [Vitis vinifera] |
| 10 |
Hb_179520_010 |
0.2240080331 |
- |
- |
PREDICTED: uncharacterized protein LOC105763691 [Gossypium raimondii] |
| 11 |
Hb_004064_020 |
0.2264867513 |
- |
- |
hypothetical protein M569_06853, partial [Genlisea aurea] |
| 12 |
Hb_143766_100 |
0.2286437495 |
- |
- |
PREDICTED: uncharacterized protein LOC105637800 [Jatropha curcas] |
| 13 |
Hb_004291_010 |
0.2303194845 |
- |
- |
(R)-limonene synthase, putative [Ricinus communis] |
| 14 |
Hb_000161_150 |
0.2313908347 |
- |
- |
PREDICTED: protein argonaute 2-like [Jatropha curcas] |
| 15 |
Hb_001258_140 |
0.2361433751 |
- |
- |
- |
| 16 |
Hb_001579_140 |
0.2440132463 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_081504_010 |
0.2447065223 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 18 |
Hb_001959_130 |
0.2477086458 |
- |
- |
transporter, putative [Ricinus communis] |
| 19 |
Hb_003052_210 |
0.2519817605 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_000720_190 |
0.2527521464 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |