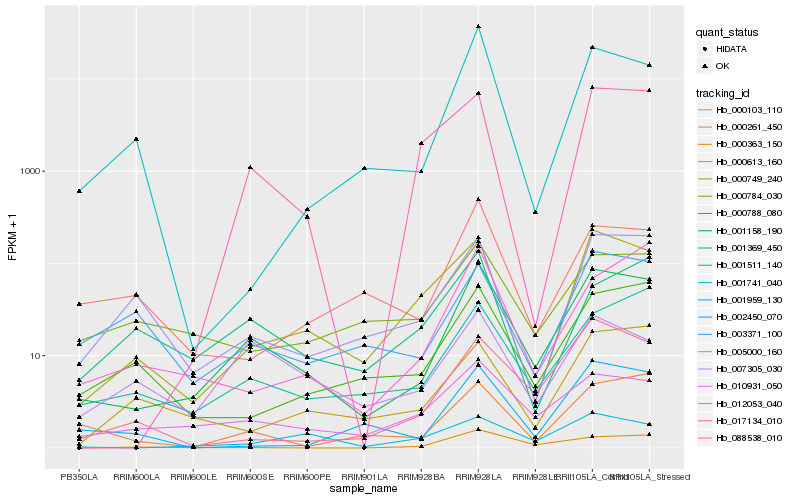
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001158_190 |
0.0 |
- |
- |
PREDICTED: bidirectional sugar transporter SWEET10-like [Jatropha curcas] |
| 2 |
Hb_010931_050 |
0.1786759601 |
transcription factor |
TF Family: Jumonji |
PREDICTED: lysine-specific demethylase JMJ25-like [Jatropha curcas] |
| 3 |
Hb_000749_240 |
0.1848232189 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001511_140 |
0.192451964 |
- |
- |
PREDICTED: RING-H2 finger protein ATL66 [Jatropha curcas] |
| 5 |
Hb_088538_010 |
0.2077020135 |
- |
- |
hypothetical protein JCGZ_12177 [Jatropha curcas] |
| 6 |
Hb_000788_080 |
0.2081782342 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 7 |
Hb_000261_450 |
0.210215577 |
- |
- |
Pectinesterase U1 precursor, putative [Ricinus communis] |
| 8 |
Hb_000363_150 |
0.2107357306 |
- |
- |
hypothetical protein POPTR_0011s09040g [Populus trichocarpa] |
| 9 |
Hb_005000_160 |
0.2110546826 |
- |
- |
PREDICTED: uncharacterized protein LOC105637882 [Jatropha curcas] |
| 10 |
Hb_017134_010 |
0.2138185167 |
- |
- |
HEV1.2 [Hevea brasiliensis] |
| 11 |
Hb_000784_030 |
0.2167261392 |
- |
- |
neutral/alkaline invertase 2 [Hevea brasiliensis] |
| 12 |
Hb_003371_100 |
0.2176710696 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 13 |
Hb_001959_130 |
0.2183249353 |
- |
- |
transporter, putative [Ricinus communis] |
| 14 |
Hb_012053_040 |
0.2196400402 |
- |
- |
PREDICTED: putative phospholipid:diacylglycerol acyltransferase 2 [Jatropha curcas] |
| 15 |
Hb_000103_110 |
0.2198405835 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_007305_030 |
0.2199779437 |
- |
- |
serine-threonine protein kinase, putative [Ricinus communis] |
| 17 |
Hb_002450_070 |
0.2215485451 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 18 |
Hb_001741_040 |
0.2249735227 |
rubber biosynthesis |
Gene Name: REF3 |
REF-like stress related protein 1 [Hevea brasiliensis] |
| 19 |
Hb_000613_160 |
0.2274252614 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 20 |
Hb_001369_450 |
0.231614388 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |