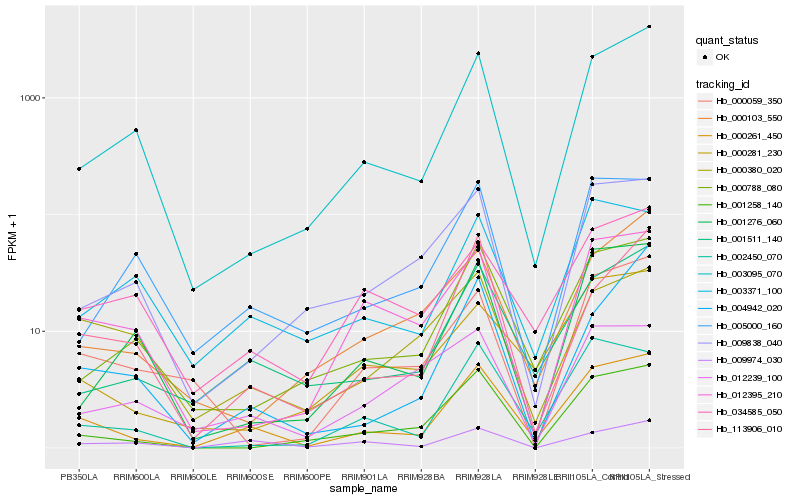
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_450 |
0.0 |
- |
- |
Pectinesterase U1 precursor, putative [Ricinus communis] |
| 2 |
Hb_001511_140 |
0.1481161709 |
- |
- |
PREDICTED: RING-H2 finger protein ATL66 [Jatropha curcas] |
| 3 |
Hb_003095_070 |
0.1664335573 |
- |
- |
hypothetical protein POPTR_0009s11230g [Populus trichocarpa] |
| 4 |
Hb_000788_080 |
0.1688119629 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 5 |
Hb_002450_070 |
0.1737657068 |
- |
- |
(S)-N-methylcoclaurine 3'-hydroxylase isozyme, putative [Ricinus communis] |
| 6 |
Hb_001258_140 |
0.1817430896 |
- |
- |
- |
| 7 |
Hb_000281_230 |
0.1835714284 |
- |
- |
PREDICTED: probable lysine--tRNA ligase, cytoplasmic isoform X2 [Jatropha curcas] |
| 8 |
Hb_003371_100 |
0.1894619538 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 9 |
Hb_009974_030 |
0.1899339773 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g18390, mitochondrial-like [Jatropha curcas] |
| 10 |
Hb_113906_010 |
0.1911532693 |
- |
- |
hypothetical protein JCGZ_18857 [Jatropha curcas] |
| 11 |
Hb_009838_040 |
0.1912310671 |
- |
- |
hypothetical protein JCGZ_12866 [Jatropha curcas] |
| 12 |
Hb_012395_210 |
0.1915296178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_034585_050 |
0.1927079014 |
- |
- |
- |
| 14 |
Hb_005000_160 |
0.192884969 |
- |
- |
PREDICTED: uncharacterized protein LOC105637882 [Jatropha curcas] |
| 15 |
Hb_001276_060 |
0.1982563425 |
- |
- |
hypothetical protein POPTR_0015s00420g [Populus trichocarpa] |
| 16 |
Hb_000103_550 |
0.1986366317 |
- |
- |
PREDICTED: guanine nucleotide-binding protein subunit gamma 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000059_350 |
0.2035456386 |
- |
- |
glutamate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_000380_020 |
0.2086916189 |
transcription factor |
TF Family: C3H |
PREDICTED: zinc finger protein 36, C3H1 type-like 3 [Jatropha curcas] |
| 19 |
Hb_012239_100 |
0.2090151354 |
- |
- |
hypothetical protein VITISV_043897 [Vitis vinifera] |
| 20 |
Hb_004942_020 |
0.2091142473 |
- |
- |
Potassium transporter, putative [Ricinus communis] |