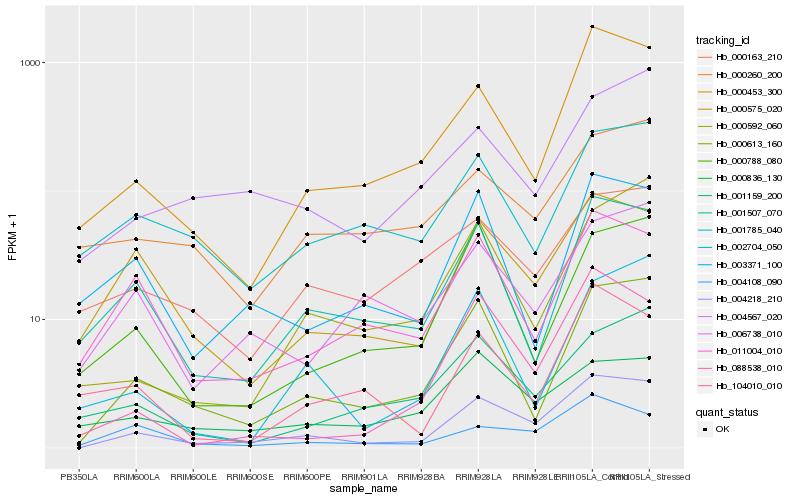
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004218_210 |
0.0 |
transcription factor |
TF Family: Orphans |
hypothetical protein JCGZ_24525 [Jatropha curcas] |
| 2 |
Hb_000613_160 |
0.1573349813 |
- |
- |
coated vesicle membrane protein, putative [Ricinus communis] |
| 3 |
Hb_000453_300 |
0.1622131242 |
- |
- |
latex cystatin [Hevea brasiliensis] |
| 4 |
Hb_000575_020 |
0.1732845047 |
- |
- |
PREDICTED: copper-transporting ATPase PAA1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_088538_010 |
0.1860234469 |
- |
- |
hypothetical protein JCGZ_12177 [Jatropha curcas] |
| 6 |
Hb_001507_070 |
0.1865016304 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_011004_010 |
0.1930359417 |
- |
- |
Mitochondrial carnitine/acylcarnitine carrier protein, putative [Ricinus communis] |
| 8 |
Hb_004108_090 |
0.196894307 |
- |
- |
PREDICTED: uncharacterized protein LOC105650231 [Jatropha curcas] |
| 9 |
Hb_006738_010 |
0.1991892284 |
- |
- |
PREDICTED: uncharacterized protein LOC105634993 [Jatropha curcas] |
| 10 |
Hb_000788_080 |
0.1998264881 |
- |
- |
PREDICTED: uncharacterized protein LOC105629703 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001159_200 |
0.2016883537 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_002704_050 |
0.205495973 |
- |
- |
cysteine-type peptidase, putative [Ricinus communis] |
| 13 |
Hb_004567_020 |
0.2058169846 |
- |
- |
Small ubiquitin-related modifier 2 [Morus notabilis] |
| 14 |
Hb_000592_060 |
0.206395017 |
- |
- |
PREDICTED: ras-related protein RABA6a [Populus euphratica] |
| 15 |
Hb_001785_040 |
0.2067045464 |
- |
- |
cytochrome b5 isoform Cb5-A [Vernicia fordii] |
| 16 |
Hb_003371_100 |
0.2077528816 |
transcription factor |
TF Family: HB |
homeobox protein, putative [Ricinus communis] |
| 17 |
Hb_000836_130 |
0.2095341627 |
- |
- |
PREDICTED: uncharacterized protein LOC105642953 isoform X2 [Jatropha curcas] |
| 18 |
Hb_000260_200 |
0.209543926 |
- |
- |
- |
| 19 |
Hb_000163_210 |
0.2095567638 |
- |
- |
PREDICTED: NADH dehydrogenase [ubiquinone] iron-sulfur protein 5-B [Tarenaya hassleriana] |
| 20 |
Hb_104010_010 |
0.2099622881 |
- |
- |
hypothetical protein JCGZ_26423 [Jatropha curcas] |