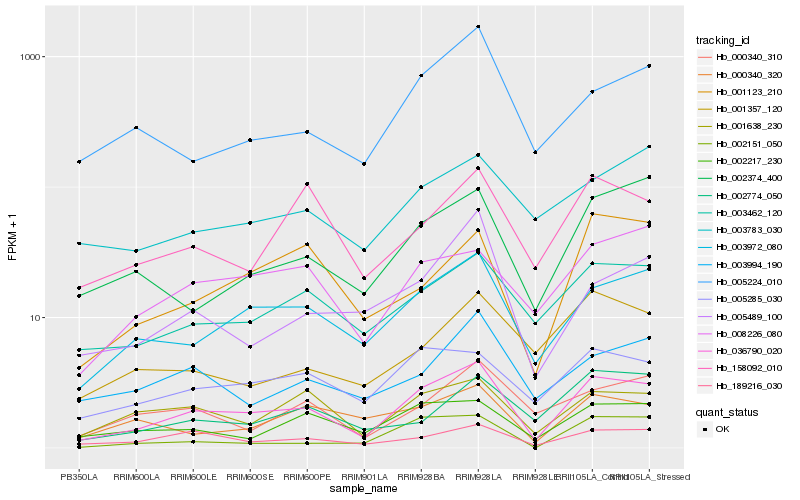
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_036790_020 |
0.0 |
- |
- |
PREDICTED: polyadenylate-binding protein 3 [Jatropha curcas] |
| 2 |
Hb_003972_080 |
0.1371201456 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 1 [Jatropha curcas] |
| 3 |
Hb_189216_030 |
0.1613044068 |
- |
- |
Pleiotropic drug resistance protein, putative [Ricinus communis] |
| 4 |
Hb_001638_230 |
0.1629864836 |
- |
- |
RNA-binding region-containing protein, putative [Ricinus communis] |
| 5 |
Hb_003462_120 |
0.168129793 |
- |
- |
Acidic endochitinase [Theobroma cacao] |
| 6 |
Hb_002217_230 |
0.1734052006 |
- |
- |
PREDICTED: uncharacterized protein LOC105643583 [Jatropha curcas] |
| 7 |
Hb_005285_030 |
0.1739918429 |
- |
- |
hypothetical protein JCGZ_20905 [Jatropha curcas] |
| 8 |
Hb_008226_080 |
0.1782004155 |
- |
- |
Protein LRP16, putative [Ricinus communis] |
| 9 |
Hb_002374_400 |
0.1789117751 |
transcription factor |
TF Family: TRAF |
protein with unknown function [Ricinus communis] |
| 10 |
Hb_005224_010 |
0.1805576124 |
- |
- |
glycine-rich protein [Bruguiera gymnorhiza] |
| 11 |
Hb_001123_210 |
0.1816482303 |
- |
- |
PREDICTED: uncharacterized protein LOC105647324 [Jatropha curcas] |
| 12 |
Hb_158092_010 |
0.1878754472 |
- |
- |
CYP51 [Hevea brasiliensis] |
| 13 |
Hb_000340_310 |
0.1880222798 |
- |
- |
PREDICTED: putative E3 ubiquitin-protein ligase RF298 [Jatropha curcas] |
| 14 |
Hb_003994_190 |
0.1908928426 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_005489_100 |
0.1916061623 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_000340_320 |
0.1936158731 |
- |
- |
PREDICTED: uncharacterized protein LOC105650052 [Jatropha curcas] |
| 17 |
Hb_002151_050 |
0.1956976794 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 18 |
Hb_003783_030 |
0.1991021135 |
- |
- |
hypothetical protein POPTR_0018s13980g [Populus trichocarpa] |
| 19 |
Hb_001357_120 |
0.1998045062 |
- |
- |
ubiquitin-activating enzyme E1, putative [Ricinus communis] |
| 20 |
Hb_002774_050 |
0.1998047067 |
- |
- |
PREDICTED: uncharacterized protein LOC105630601 [Jatropha curcas] |