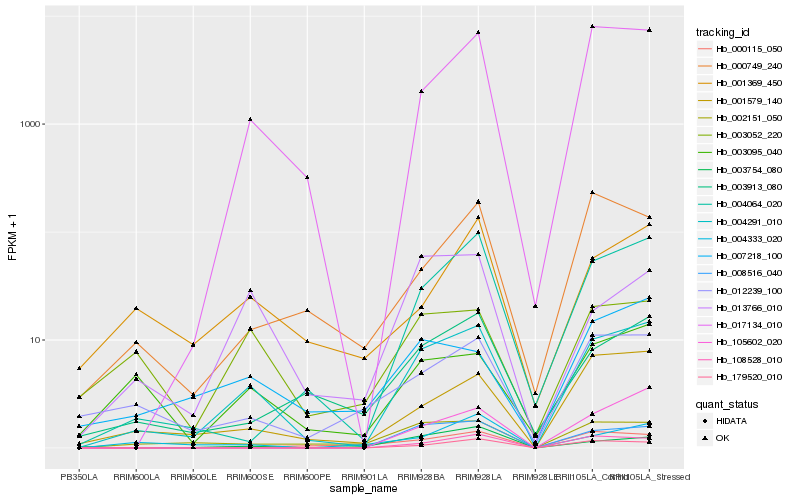
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004291_010 |
0.0 |
- |
- |
(R)-limonene synthase, putative [Ricinus communis] |
| 2 |
Hb_017134_010 |
0.1715944703 |
- |
- |
HEV1.2 [Hevea brasiliensis] |
| 3 |
Hb_001579_140 |
0.2004606471 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X2 [Jatropha curcas] |
| 4 |
Hb_004064_020 |
0.2031394918 |
- |
- |
hypothetical protein M569_06853, partial [Genlisea aurea] |
| 5 |
Hb_002151_050 |
0.2083764834 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 6 |
Hb_003913_080 |
0.2099348765 |
- |
- |
rpl27A [Hemiselmis andersenii] |
| 7 |
Hb_012239_100 |
0.2215879403 |
- |
- |
hypothetical protein VITISV_043897 [Vitis vinifera] |
| 8 |
Hb_013766_010 |
0.2236087273 |
- |
- |
hypothetical protein JCGZ_00483 [Jatropha curcas] |
| 9 |
Hb_003095_040 |
0.2247078677 |
- |
- |
hypothetical protein CICLE_v10030016mg [Citrus clementina] |
| 10 |
Hb_003052_220 |
0.2293347735 |
- |
- |
- |
| 11 |
Hb_008516_040 |
0.2302952706 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000115_050 |
0.2303194845 |
- |
- |
- |
| 13 |
Hb_007218_100 |
0.2333520845 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 14 |
Hb_108528_010 |
0.2394283785 |
- |
- |
unnamed protein product [Coffea canephora] |
| 15 |
Hb_001369_450 |
0.2395377754 |
- |
- |
trehalose-6-phosphate synthase, putative [Ricinus communis] |
| 16 |
Hb_004333_020 |
0.2407057095 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 17 |
Hb_000749_240 |
0.2411632469 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_105602_020 |
0.2418384335 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1-like [Populus euphratica] |
| 19 |
Hb_003754_080 |
0.2441320906 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |
| 20 |
Hb_179520_010 |
0.2466869619 |
- |
- |
PREDICTED: uncharacterized protein LOC105763691 [Gossypium raimondii] |