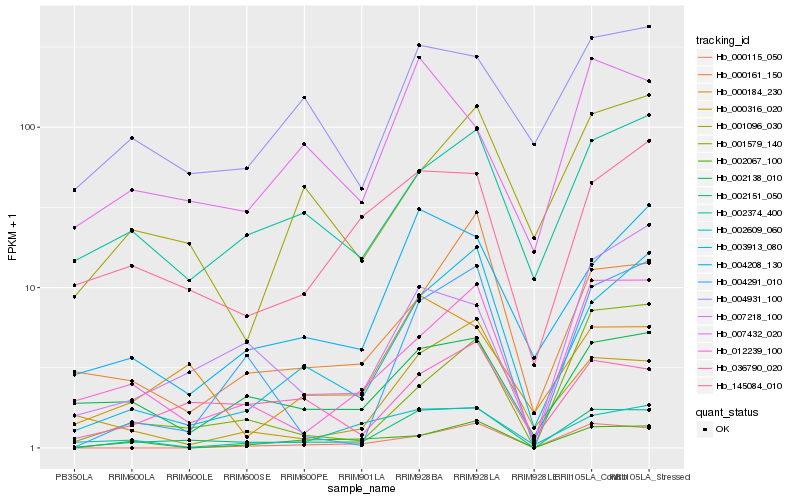
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002151_050 |
0.0 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 2 |
Hb_003913_080 |
0.1879444131 |
- |
- |
rpl27A [Hemiselmis andersenii] |
| 3 |
Hb_012239_100 |
0.1888636051 |
- |
- |
hypothetical protein VITISV_043897 [Vitis vinifera] |
| 4 |
Hb_002138_010 |
0.1949561552 |
- |
- |
hypothetical protein POPTR_0001s30620g [Populus trichocarpa] |
| 5 |
Hb_036790_020 |
0.1956976794 |
- |
- |
PREDICTED: polyadenylate-binding protein 3 [Jatropha curcas] |
| 6 |
Hb_000184_230 |
0.2009473197 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein CONSTANS-LIKE 9-like [Jatropha curcas] |
| 7 |
Hb_145084_010 |
0.2012765435 |
- |
- |
PREDICTED: uncharacterized protein LOC105646270 [Jatropha curcas] |
| 8 |
Hb_004208_130 |
0.2016923497 |
- |
- |
hypothetical protein JCGZ_16532 [Jatropha curcas] |
| 9 |
Hb_004291_010 |
0.2083764834 |
- |
- |
(R)-limonene synthase, putative [Ricinus communis] |
| 10 |
Hb_002374_400 |
0.2122882332 |
transcription factor |
TF Family: TRAF |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_007218_100 |
0.2126643795 |
- |
- |
phosphofructokinase [Hevea brasiliensis] |
| 12 |
Hb_000115_050 |
0.2131743935 |
- |
- |
- |
| 13 |
Hb_007432_020 |
0.2182767085 |
- |
- |
hypothetical protein POPTR_0010s13310g [Populus trichocarpa] |
| 14 |
Hb_004931_100 |
0.2204397477 |
- |
- |
Actin depolymerizing factor 6 isoform 1 [Theobroma cacao] |
| 15 |
Hb_002609_060 |
0.2213162883 |
- |
- |
PREDICTED: sugar transport protein 8-like [Jatropha curcas] |
| 16 |
Hb_000161_150 |
0.2231679684 |
- |
- |
PREDICTED: protein argonaute 2-like [Jatropha curcas] |
| 17 |
Hb_000316_020 |
0.2283153661 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 18 |
Hb_001096_030 |
0.2313041618 |
- |
- |
uridylate kinase plant, putative [Ricinus communis] |
| 19 |
Hb_002067_100 |
0.231535384 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001579_140 |
0.2332890473 |
- |
- |
PREDICTED: probable inactive poly [ADP-ribose] polymerase SRO2 isoform X2 [Jatropha curcas] |