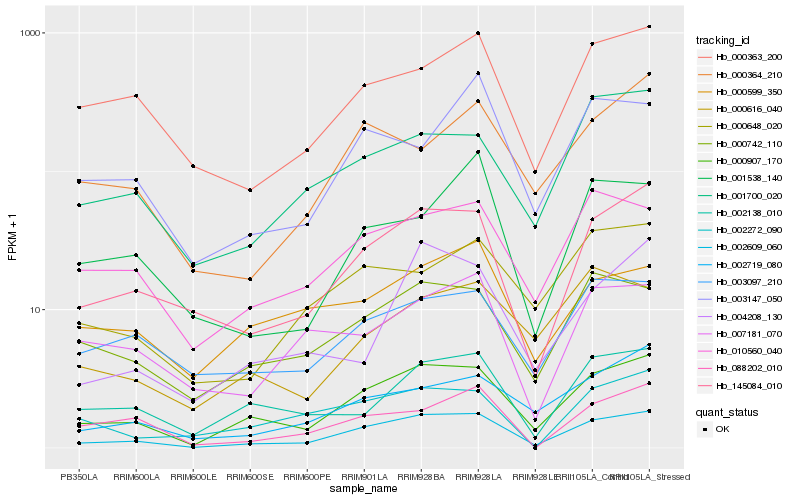
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002609_060 |
0.0 |
- |
- |
PREDICTED: sugar transport protein 8-like [Jatropha curcas] |
| 2 |
Hb_000907_170 |
0.0780402576 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 3 |
Hb_145084_010 |
0.1242157285 |
- |
- |
PREDICTED: uncharacterized protein LOC105646270 [Jatropha curcas] |
| 4 |
Hb_000742_110 |
0.1601616423 |
- |
- |
hypothetical protein JCGZ_09215 [Jatropha curcas] |
| 5 |
Hb_010560_040 |
0.1653657989 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 11 [Jatropha curcas] |
| 6 |
Hb_000363_200 |
0.1710755372 |
- |
- |
40S ribosomal protein S8, putative [Ricinus communis] |
| 7 |
Hb_002719_080 |
0.173071482 |
- |
- |
hypothetical protein POPTR_0006s27920g [Populus trichocarpa] |
| 8 |
Hb_002138_010 |
0.1754333696 |
- |
- |
hypothetical protein POPTR_0001s30620g [Populus trichocarpa] |
| 9 |
Hb_001700_020 |
0.1779358478 |
- |
- |
PREDICTED: 60S ribosomal protein L37-1 [Jatropha curcas] |
| 10 |
Hb_088202_010 |
0.179814337 |
- |
- |
hypothetical protein JCGZ_13241 [Jatropha curcas] |
| 11 |
Hb_003097_210 |
0.1801357817 |
- |
- |
PREDICTED: uncharacterized protein LOC105631194 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000648_020 |
0.1813580121 |
- |
- |
PREDICTED: chaperone protein dnaJ 10-like [Jatropha curcas] |
| 13 |
Hb_001538_140 |
0.1826411322 |
- |
- |
PREDICTED: PXMP2/4 family protein 4-like [Jatropha curcas] |
| 14 |
Hb_004208_130 |
0.1848111166 |
- |
- |
hypothetical protein JCGZ_16532 [Jatropha curcas] |
| 15 |
Hb_003147_050 |
0.186778966 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 16 |
Hb_000364_210 |
0.1873741518 |
- |
- |
Mitochondrial import receptor subunit TOM7-1, putative [Ricinus communis] |
| 17 |
Hb_007181_070 |
0.1885324518 |
- |
- |
PREDICTED: alpha-galactosidase-like [Jatropha curcas] |
| 18 |
Hb_002272_090 |
0.1894456681 |
- |
- |
RING/U-box superfamily protein [Theobroma cacao] |
| 19 |
Hb_000616_040 |
0.1915685962 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 20 |
Hb_000599_350 |
0.1934332114 |
- |
- |
PREDICTED: glutathione gamma-glutamylcysteinyltransferase 1 [Jatropha curcas] |