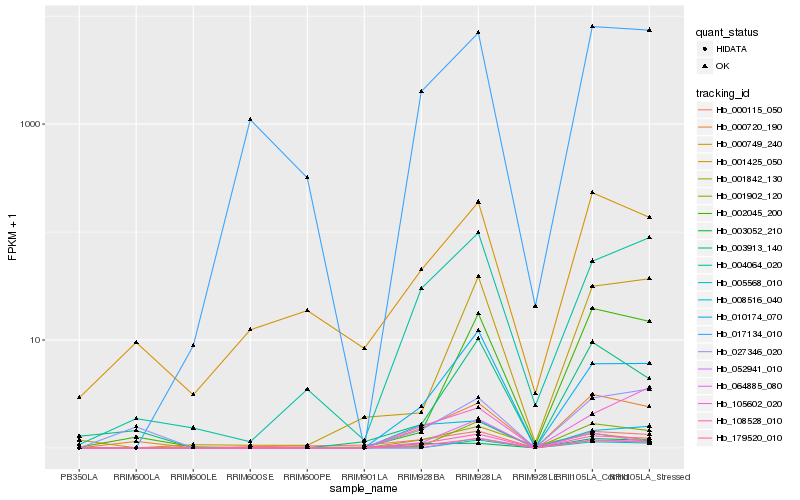
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_108528_010 |
0.0 |
- |
- |
unnamed protein product [Coffea canephora] |
| 2 |
Hb_179520_010 |
0.0228842723 |
- |
- |
PREDICTED: uncharacterized protein LOC105763691 [Gossypium raimondii] |
| 3 |
Hb_010174_070 |
0.1315457185 |
- |
- |
- |
| 4 |
Hb_000720_190 |
0.1385814534 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 5 |
Hb_004064_020 |
0.1545628244 |
- |
- |
hypothetical protein M569_06853, partial [Genlisea aurea] |
| 6 |
Hb_001902_120 |
0.1640057138 |
- |
- |
PREDICTED: uncharacterized protein LOC105648455 isoform X2 [Jatropha curcas] |
| 7 |
Hb_003052_210 |
0.1749630917 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_008516_040 |
0.1870262181 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_017134_010 |
0.187977921 |
- |
- |
HEV1.2 [Hevea brasiliensis] |
| 10 |
Hb_001842_130 |
0.1979895877 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_002045_200 |
0.2116432869 |
- |
- |
PREDICTED: beta-carotene isomerase D27, chloroplastic [Jatropha curcas] |
| 12 |
Hb_105602_020 |
0.2124083666 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1-like [Populus euphratica] |
| 13 |
Hb_001425_050 |
0.2154582667 |
- |
- |
- |
| 14 |
Hb_000749_240 |
0.217321527 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000115_050 |
0.218385863 |
- |
- |
- |
| 16 |
Hb_003913_140 |
0.2204297132 |
- |
- |
PREDICTED: uncharacterized protein LOC105640324 [Jatropha curcas] |
| 17 |
Hb_064885_080 |
0.2212351712 |
- |
- |
- |
| 18 |
Hb_027346_020 |
0.2358190664 |
- |
- |
- |
| 19 |
Hb_052941_010 |
0.2367206765 |
- |
- |
PREDICTED: probable peroxidase 61 [Jatropha curcas] |
| 20 |
Hb_005568_010 |
0.2374229008 |
- |
- |
- |