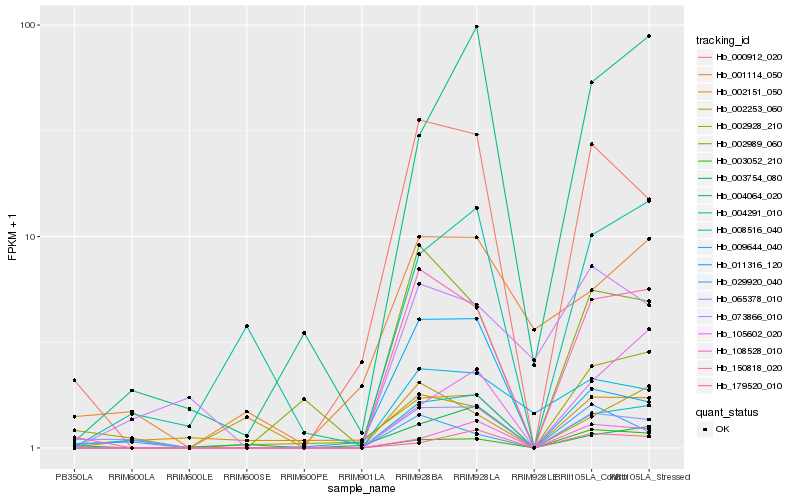
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_150818_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645039 [Jatropha curcas] |
| 2 |
Hb_008516_040 |
0.1347041948 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_002989_060 |
0.1603839307 |
- |
- |
hypothetical protein JCGZ_22461 [Jatropha curcas] |
| 4 |
Hb_000912_020 |
0.1641533516 |
- |
- |
PREDICTED: uncharacterized protein LOC105645039 [Jatropha curcas] |
| 5 |
Hb_003052_210 |
0.2019666791 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_065378_010 |
0.2168102688 |
- |
- |
S-locus-specific glycoprotein S13 precursor, putative [Ricinus communis] |
| 7 |
Hb_009644_040 |
0.2396507221 |
- |
- |
PREDICTED: protein SKIP34 [Jatropha curcas] |
| 8 |
Hb_108528_010 |
0.258392584 |
- |
- |
unnamed protein product [Coffea canephora] |
| 9 |
Hb_004291_010 |
0.2663809733 |
- |
- |
(R)-limonene synthase, putative [Ricinus communis] |
| 10 |
Hb_105602_020 |
0.2699623155 |
- |
- |
PREDICTED: 60S ribosomal protein L19-1-like [Populus euphratica] |
| 11 |
Hb_029920_040 |
0.2715620306 |
- |
- |
PREDICTED: uncharacterized protein LOC105635837 [Jatropha curcas] |
| 12 |
Hb_179520_010 |
0.2755540265 |
- |
- |
PREDICTED: uncharacterized protein LOC105763691 [Gossypium raimondii] |
| 13 |
Hb_004064_020 |
0.2761789869 |
- |
- |
hypothetical protein M569_06853, partial [Genlisea aurea] |
| 14 |
Hb_002928_210 |
0.2773347428 |
- |
- |
- |
| 15 |
Hb_002151_050 |
0.2804766906 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 16 |
Hb_002253_060 |
0.2811084755 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_001114_050 |
0.2811751846 |
- |
- |
hypothetical protein RCOM_0744580 [Ricinus communis] |
| 18 |
Hb_011316_120 |
0.2845195419 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor bHLH118-like [Jatropha curcas] |
| 19 |
Hb_073866_010 |
0.2852001053 |
- |
- |
PREDICTED: N-alpha-acetyltransferase 20-like [Jatropha curcas] |
| 20 |
Hb_003754_080 |
0.2903400865 |
- |
- |
PREDICTED: uncharacterized protein LOC105634374 [Jatropha curcas] |