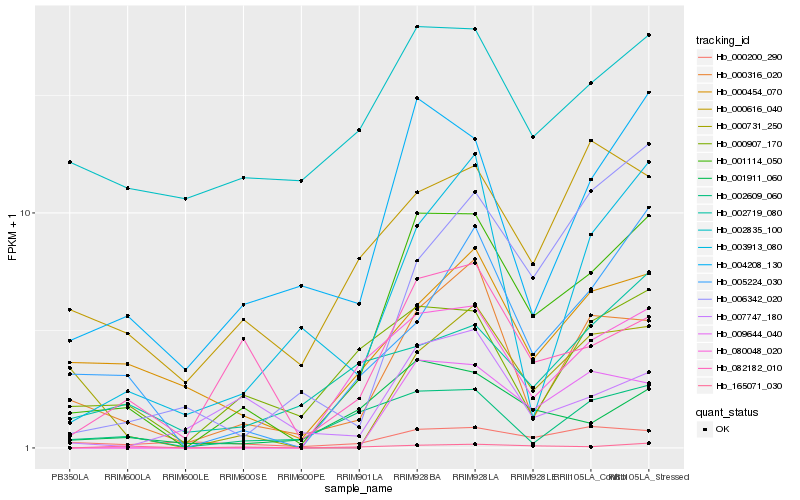
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001114_050 |
0.0 |
- |
- |
hypothetical protein RCOM_0744580 [Ricinus communis] |
| 2 |
Hb_004208_130 |
0.1953464902 |
- |
- |
hypothetical protein JCGZ_16532 [Jatropha curcas] |
| 3 |
Hb_080048_020 |
0.198285792 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000316_020 |
0.198576335 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 5 |
Hb_001911_060 |
0.2122112206 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_009644_040 |
0.2172613733 |
- |
- |
PREDICTED: protein SKIP34 [Jatropha curcas] |
| 7 |
Hb_082182_010 |
0.2194748725 |
- |
- |
PREDICTED: protein SUPPRESSOR OF npr1-1, CONSTITUTIVE 1-like [Jatropha curcas] |
| 8 |
Hb_005224_030 |
0.2197007411 |
- |
- |
Dipeptide transport ATP-binding protein dppF [Theobroma cacao] |
| 9 |
Hb_002609_060 |
0.2393496067 |
- |
- |
PREDICTED: sugar transport protein 8-like [Jatropha curcas] |
| 10 |
Hb_000907_170 |
0.2404288703 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 11 |
Hb_000454_070 |
0.2417371985 |
- |
- |
PREDICTED: 30S ribosomal protein S31, mitochondrial [Cucumis sativus] |
| 12 |
Hb_000616_040 |
0.2445784489 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 13 |
Hb_003913_080 |
0.2473668925 |
- |
- |
rpl27A [Hemiselmis andersenii] |
| 14 |
Hb_000731_250 |
0.2508226903 |
- |
- |
- |
| 15 |
Hb_006342_020 |
0.2513202021 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 16 |
Hb_007747_180 |
0.2533975788 |
transcription factor |
TF Family: TCP |
PREDICTED: bifunctional nitrilase/nitrile hydratase NIT4B-like [Jatropha curcas] |
| 17 |
Hb_000200_290 |
0.2567730806 |
- |
- |
PREDICTED: elongator complex protein 2 isoform X1 [Jatropha curcas] |
| 18 |
Hb_165071_030 |
0.2582204804 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_002719_080 |
0.260816043 |
- |
- |
hypothetical protein POPTR_0006s27920g [Populus trichocarpa] |
| 20 |
Hb_002835_100 |
0.2621196149 |
- |
- |
PREDICTED: protein DEHYDRATION-INDUCED 19 homolog 6-like [Jatropha curcas] |