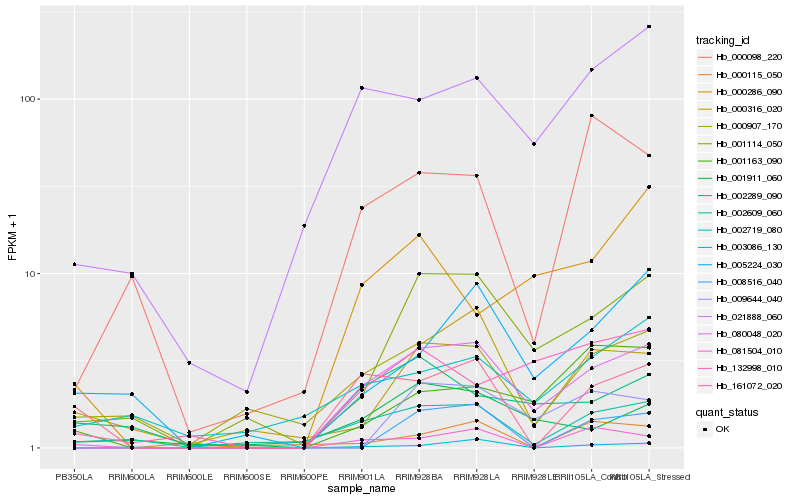
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_080048_020 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001114_050 |
0.198285792 |
- |
- |
hypothetical protein RCOM_0744580 [Ricinus communis] |
| 3 |
Hb_021888_060 |
0.2214635622 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 15 homolog [Jatropha curcas] |
| 4 |
Hb_002609_060 |
0.2452848966 |
- |
- |
PREDICTED: sugar transport protein 8-like [Jatropha curcas] |
| 5 |
Hb_009644_040 |
0.2548630776 |
- |
- |
PREDICTED: protein SKIP34 [Jatropha curcas] |
| 6 |
Hb_132998_010 |
0.256522573 |
- |
- |
hypothetical protein JCGZ_00197 [Jatropha curcas] |
| 7 |
Hb_001911_060 |
0.2566986477 |
desease resistance |
Gene Name: NB-ARC |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_000286_090 |
0.2704019605 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_00197 [Jatropha curcas] |
| 9 |
Hb_000098_220 |
0.2705510866 |
- |
- |
PREDICTED: VQ motif-containing protein 4 [Jatropha curcas] |
| 10 |
Hb_001163_090 |
0.2729059769 |
- |
- |
hypothetical protein POPTR_0009s14920g [Populus trichocarpa] |
| 11 |
Hb_000907_170 |
0.2732907419 |
- |
- |
leucine-rich repeat-containing protein, putative [Ricinus communis] |
| 12 |
Hb_161072_020 |
0.2772003142 |
- |
- |
- |
| 13 |
Hb_002289_090 |
0.2773052248 |
- |
- |
cytoplasmic tRNA 2-thiolation protein 2-like [Scleropages formosus] |
| 14 |
Hb_081504_010 |
0.2823189494 |
- |
- |
PREDICTED: uncharacterized protein LOC105795045 [Gossypium raimondii] |
| 15 |
Hb_003086_130 |
0.2823729483 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 16 |
Hb_005224_030 |
0.2857058515 |
- |
- |
Dipeptide transport ATP-binding protein dppF [Theobroma cacao] |
| 17 |
Hb_008516_040 |
0.286494596 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_000316_020 |
0.2867192597 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 19 |
Hb_002719_080 |
0.2874578764 |
- |
- |
hypothetical protein POPTR_0006s27920g [Populus trichocarpa] |
| 20 |
Hb_000115_050 |
0.288927385 |
- |
- |
- |