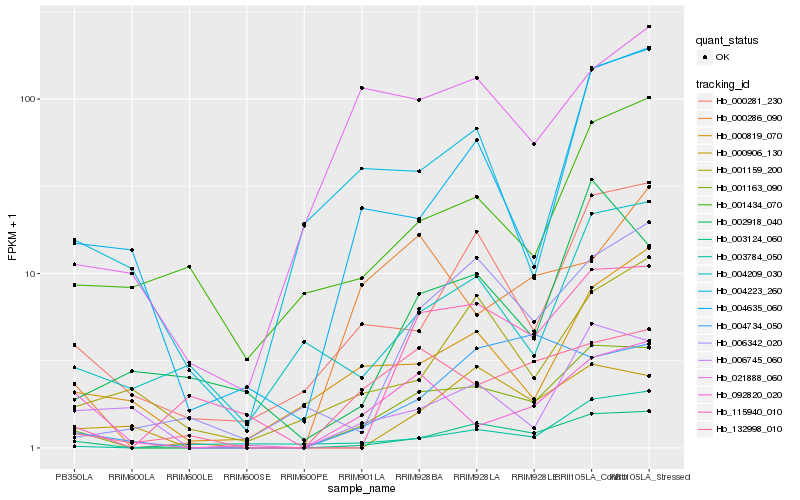
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001163_090 |
0.0 |
- |
- |
hypothetical protein POPTR_0009s14920g [Populus trichocarpa] |
| 2 |
Hb_003124_060 |
0.1158430125 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |
| 3 |
Hb_132998_010 |
0.1960280477 |
- |
- |
hypothetical protein JCGZ_00197 [Jatropha curcas] |
| 4 |
Hb_000281_230 |
0.2043893102 |
- |
- |
PREDICTED: probable lysine--tRNA ligase, cytoplasmic isoform X2 [Jatropha curcas] |
| 5 |
Hb_115940_010 |
0.2141085742 |
- |
- |
putative auxin-amidohydrolase precursor family protein [Populus trichocarpa] |
| 6 |
Hb_006342_020 |
0.2214087049 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_004635_060 |
0.2308091475 |
- |
- |
hypothetical protein POPTR_0006s22850g [Populus trichocarpa] |
| 8 |
Hb_003784_050 |
0.2323606532 |
- |
- |
PREDICTED: replication protein A 32 kDa subunit B [Jatropha curcas] |
| 9 |
Hb_092820_020 |
0.2380688709 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_006745_060 |
0.2387264863 |
- |
- |
- |
| 11 |
Hb_021888_060 |
0.2405296879 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 15 homolog [Jatropha curcas] |
| 12 |
Hb_000286_090 |
0.2419809196 |
transcription factor |
TF Family: B3 |
hypothetical protein JCGZ_00197 [Jatropha curcas] |
| 13 |
Hb_004223_260 |
0.2445832894 |
- |
- |
hypothetical protein F383_09277 [Gossypium arboreum] |
| 14 |
Hb_000819_070 |
0.245676436 |
- |
- |
PREDICTED: dolichol-phosphate mannosyltransferase subunit 1 [Jatropha curcas] |
| 15 |
Hb_001159_200 |
0.2485022644 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_004209_030 |
0.2509413828 |
- |
- |
PREDICTED: (DL)-glycerol-3-phosphatase 2 isoform X1 [Jatropha curcas] |
| 17 |
Hb_000906_130 |
0.2524444918 |
- |
- |
- |
| 18 |
Hb_001434_070 |
0.2545866928 |
- |
- |
PREDICTED: mitochondrial import inner membrane translocase subunit TIM8 [Jatropha curcas] |
| 19 |
Hb_004734_050 |
0.2556798596 |
- |
- |
- |
| 20 |
Hb_002918_040 |
0.2562986211 |
transcription factor |
TF Family: SBP |
Squamosa promoter-binding protein, putative [Ricinus communis] |