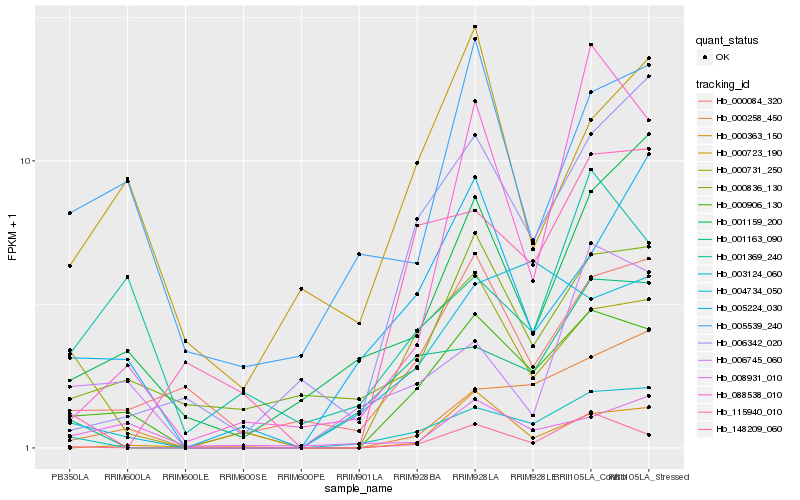
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000906_130 |
0.0 |
- |
- |
- |
| 2 |
Hb_003124_060 |
0.201779663 |
- |
- |
Beta-ureidopropionase, putative [Ricinus communis] |
| 3 |
Hb_000731_250 |
0.2036915338 |
- |
- |
- |
| 4 |
Hb_000084_320 |
0.2281734438 |
- |
- |
PREDICTED: probable small nuclear ribonucleoprotein G [Populus euphratica] |
| 5 |
Hb_088538_010 |
0.2376992295 |
- |
- |
hypothetical protein JCGZ_12177 [Jatropha curcas] |
| 6 |
Hb_005224_030 |
0.2382565744 |
- |
- |
Dipeptide transport ATP-binding protein dppF [Theobroma cacao] |
| 7 |
Hb_004734_050 |
0.2423934408 |
- |
- |
- |
| 8 |
Hb_006342_020 |
0.2428491482 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 9 |
Hb_000836_130 |
0.2437005471 |
- |
- |
PREDICTED: uncharacterized protein LOC105642953 isoform X2 [Jatropha curcas] |
| 10 |
Hb_148209_060 |
0.244182509 |
- |
- |
PREDICTED: BURP domain-containing protein 3-like [Jatropha curcas] |
| 11 |
Hb_000258_450 |
0.2477140375 |
- |
- |
- |
| 12 |
Hb_001159_200 |
0.2498451394 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_000363_150 |
0.2518158679 |
- |
- |
hypothetical protein POPTR_0011s09040g [Populus trichocarpa] |
| 14 |
Hb_001163_090 |
0.2524444918 |
- |
- |
hypothetical protein POPTR_0009s14920g [Populus trichocarpa] |
| 15 |
Hb_008931_010 |
0.2555119712 |
- |
- |
PREDICTED: macrophage erythroblast attacher [Jatropha curcas] |
| 16 |
Hb_000723_190 |
0.2580711071 |
- |
- |
Protein kinase APK1A, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_005539_240 |
0.2590239424 |
- |
- |
hypothetical protein MIMGU_mgv1a0220462mg, partial [Erythranthe guttata] |
| 18 |
Hb_006745_060 |
0.259638737 |
- |
- |
- |
| 19 |
Hb_001369_240 |
0.2609617856 |
- |
- |
- |
| 20 |
Hb_115940_010 |
0.2612901376 |
- |
- |
putative auxin-amidohydrolase precursor family protein [Populus trichocarpa] |