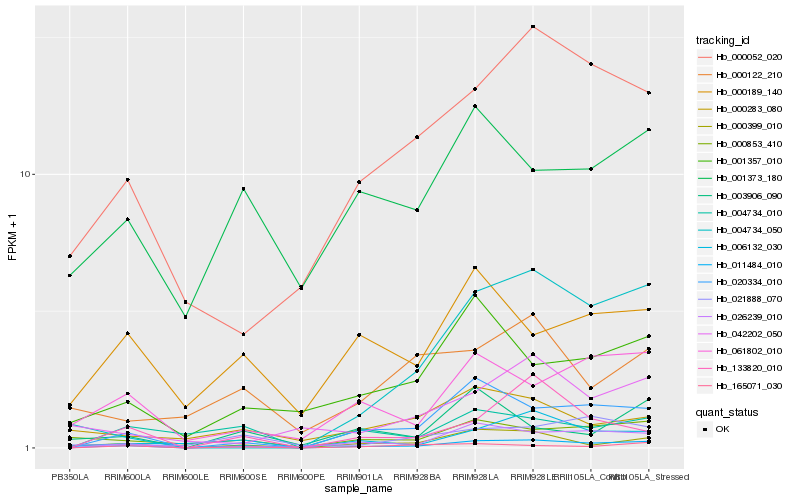
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011484_010 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 2 |
Hb_000399_010 |
0.1848811578 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 3 |
Hb_020334_010 |
0.1880907697 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 4 |
Hb_026239_010 |
0.1967799714 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 5 |
Hb_006132_030 |
0.2006913476 |
- |
- |
hypothetical protein CARUB_v10012126mg, partial [Capsella rubella] |
| 6 |
Hb_000853_410 |
0.2090013313 |
- |
- |
- |
| 7 |
Hb_061802_010 |
0.2158123523 |
- |
- |
hypothetical protein VITISV_011986 [Vitis vinifera] |
| 8 |
Hb_000052_020 |
0.2223670881 |
desease resistance |
Gene Name: ABC_tran |
PREDICTED: LOW QUALITY PROTEIN: ABC transporter G family member 2-like [Jatropha curcas] |
| 9 |
Hb_001357_010 |
0.2276178691 |
- |
- |
- |
| 10 |
Hb_021888_070 |
0.2352500518 |
- |
- |
Putative CCR4-associated factor 1 like 11 [Glycine soja] |
| 11 |
Hb_165071_030 |
0.2371505433 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 12 |
Hb_004734_050 |
0.2385358386 |
- |
- |
- |
| 13 |
Hb_133820_010 |
0.2391624037 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 14 |
Hb_000189_140 |
0.2440445295 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_001373_180 |
0.2465707385 |
- |
- |
PREDICTED: uncharacterized protein LOC105628780 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000122_210 |
0.2506864405 |
- |
- |
- |
| 17 |
Hb_000283_080 |
0.2508764929 |
- |
- |
- |
| 18 |
Hb_004734_010 |
0.2512043245 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 19 |
Hb_003906_090 |
0.2515216804 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g38010 [Jatropha curcas] |
| 20 |
Hb_042202_050 |
0.256231155 |
- |
- |
TPA: hypothetical protein ZEAMMB73_683745 [Zea mays] |