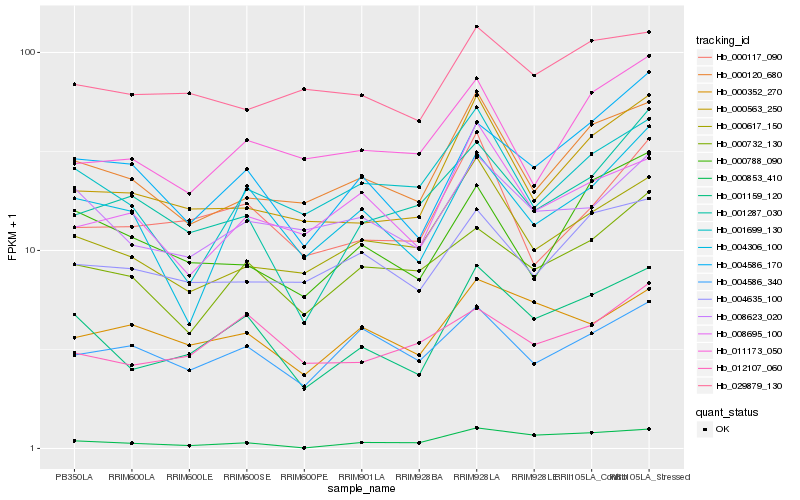
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001159_120 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645945 [Jatropha curcas] |
| 2 |
Hb_004586_170 |
0.1203140529 |
- |
- |
hypothetical protein PRUPE_ppa011196mg [Prunus persica] |
| 3 |
Hb_008623_020 |
0.1228721951 |
- |
- |
PREDICTED: uncharacterized protein At3g17611 [Jatropha curcas] |
| 4 |
Hb_000120_680 |
0.1240395054 |
- |
- |
PREDICTED: uncharacterized protein LOC105628572 [Jatropha curcas] |
| 5 |
Hb_000563_250 |
0.1244887278 |
- |
- |
PREDICTED: uncharacterized protein LOC105643472 [Jatropha curcas] |
| 6 |
Hb_000617_150 |
0.1331561838 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g61360-like [Jatropha curcas] |
| 7 |
Hb_004306_100 |
0.1365733314 |
- |
- |
PREDICTED: uncharacterized protein LOC105644390 [Jatropha curcas] |
| 8 |
Hb_000732_130 |
0.1373294634 |
transcription factor |
TF Family: HSF |
hypothetical protein JCGZ_17108 [Jatropha curcas] |
| 9 |
Hb_012107_060 |
0.1401935914 |
- |
- |
hypothetical protein JCGZ_17178 [Jatropha curcas] |
| 10 |
Hb_004635_100 |
0.1407990506 |
- |
- |
PREDICTED: L-aminoadipate-semialdehyde dehydrogenase-phosphopantetheinyl transferase isoform X1 [Jatropha curcas] |
| 11 |
Hb_000352_270 |
0.1410088816 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g26630, chloroplastic-like [Jatropha curcas] |
| 12 |
Hb_000853_410 |
0.1416655054 |
- |
- |
- |
| 13 |
Hb_000788_090 |
0.141969034 |
transcription factor |
TF Family: FAR1 |
PREDICTED: protein FAR1-RELATED SEQUENCE 11-like isoform X2 [Glycine max] |
| 14 |
Hb_001699_130 |
0.1431047281 |
transcription factor |
TF Family: CSD |
PREDICTED: cold shock domain-containing protein 3 [Jatropha curcas] |
| 15 |
Hb_000117_090 |
0.1436636879 |
- |
- |
PREDICTED: uncharacterized protein LOC105633456 [Jatropha curcas] |
| 16 |
Hb_008695_100 |
0.1471631046 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 1 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001287_030 |
0.147331632 |
- |
- |
30S ribosomal protein S31, mitochondrial precursor, putative [Ricinus communis] |
| 18 |
Hb_029879_130 |
0.1476212813 |
- |
- |
Derlin-2, putative [Ricinus communis] |
| 19 |
Hb_004586_340 |
0.1520836743 |
- |
- |
PREDICTED: serine/threonine-protein kinase PEPKR2 [Jatropha curcas] |
| 20 |
Hb_011173_050 |
0.1523280229 |
- |
- |
PREDICTED: OTU domain-containing protein DDB_G0284757 [Jatropha curcas] |