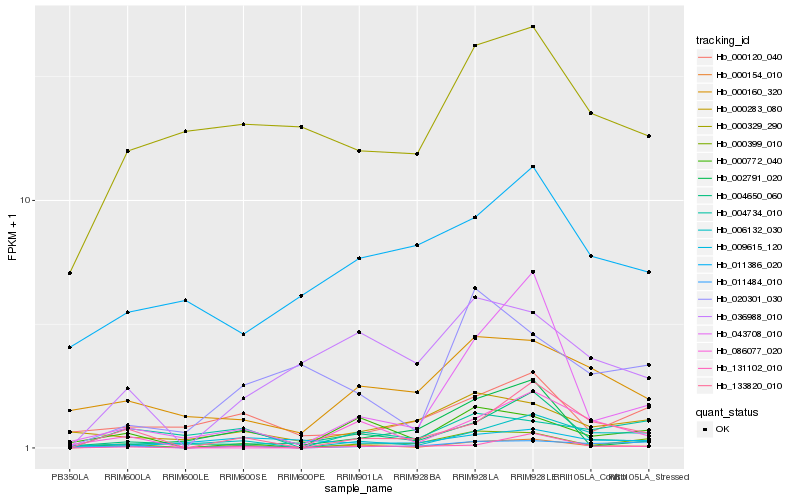
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000154_010 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_004650_060 |
0.1978366362 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |
| 3 |
Hb_133820_010 |
0.2221052207 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 4 |
Hb_006132_030 |
0.2243330587 |
- |
- |
hypothetical protein CARUB_v10012126mg, partial [Capsella rubella] |
| 5 |
Hb_043708_010 |
0.2353219639 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 6 |
Hb_000160_320 |
0.2511124593 |
- |
- |
hypothetical protein POPTR_0004s08730g [Populus trichocarpa] |
| 7 |
Hb_036988_010 |
0.2532512706 |
- |
- |
hypothetical protein JCGZ_25496 [Jatropha curcas] |
| 8 |
Hb_000772_040 |
0.2533488336 |
- |
- |
PREDICTED: uncharacterized protein LOC105036273 [Elaeis guineensis] |
| 9 |
Hb_000399_010 |
0.2625121758 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 10 |
Hb_004734_010 |
0.2629791165 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At1g12700, mitochondrial [Jatropha curcas] |
| 11 |
Hb_000329_290 |
0.2652814191 |
transcription factor |
TF Family: GRAS |
PREDICTED: LOW QUALITY PROTEIN: scarecrow-like protein 6 [Jatropha curcas] |
| 12 |
Hb_011484_010 |
0.2683166931 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 13 |
Hb_131102_010 |
0.2694398466 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_020301_030 |
0.2713502157 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 15 |
Hb_086077_020 |
0.2741991495 |
- |
- |
hypothetical protein PHAVU_001G054900g [Phaseolus vulgaris] |
| 16 |
Hb_002791_020 |
0.2765651073 |
- |
- |
hypothetical protein AALP_AA5G271700 [Arabis alpina] |
| 17 |
Hb_000120_040 |
0.2782131411 |
- |
- |
- |
| 18 |
Hb_011386_020 |
0.2799967513 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 19 |
Hb_009615_120 |
0.2800458305 |
- |
- |
gypsy/Ty-3 retroelement polyprotein; 69905-74404 [Arabidopsis thaliana] |
| 20 |
Hb_000283_080 |
0.28357033 |
- |
- |
- |