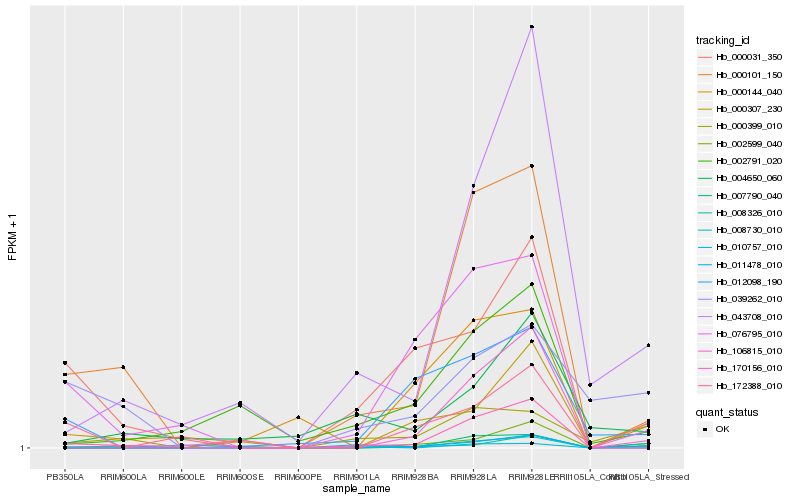
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008730_010 |
0.0 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 2 |
Hb_000101_150 |
0.162195351 |
- |
- |
- |
| 3 |
Hb_002599_040 |
0.2320331151 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 4 |
Hb_043708_010 |
0.2355436147 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 5 |
Hb_076795_010 |
0.2574210121 |
- |
- |
hypothetical protein POPTR_0001s07680g, partial [Populus trichocarpa] |
| 6 |
Hb_106815_010 |
0.2695042221 |
- |
- |
hypothetical protein, conserved [Eimeria acervulina] |
| 7 |
Hb_000399_010 |
0.2798227302 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 8 |
Hb_000307_230 |
0.2883592235 |
- |
- |
- |
| 9 |
Hb_007790_040 |
0.2953253314 |
- |
- |
PREDICTED: uncharacterized protein LOC104417177 [Eucalyptus grandis] |
| 10 |
Hb_000031_350 |
0.2965029079 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic isoform X2 [Cucumis melo] |
| 11 |
Hb_170156_010 |
0.2967490235 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 12 |
Hb_012098_190 |
0.3015310166 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 13 |
Hb_008326_010 |
0.3019865584 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 14 |
Hb_002791_020 |
0.3021083307 |
- |
- |
hypothetical protein AALP_AA5G271700 [Arabis alpina] |
| 15 |
Hb_000144_040 |
0.3028433149 |
- |
- |
hypothetical protein POPTR_0006s06790g [Populus trichocarpa] |
| 16 |
Hb_172388_010 |
0.3043048378 |
- |
- |
PREDICTED: uncharacterized protein LOC105633410 [Jatropha curcas] |
| 17 |
Hb_004650_060 |
0.3044837379 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |
| 18 |
Hb_039262_010 |
0.313009754 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 19 |
Hb_010757_010 |
0.3152101386 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 20 |
Hb_011478_010 |
0.3173160577 |
- |
- |
- |