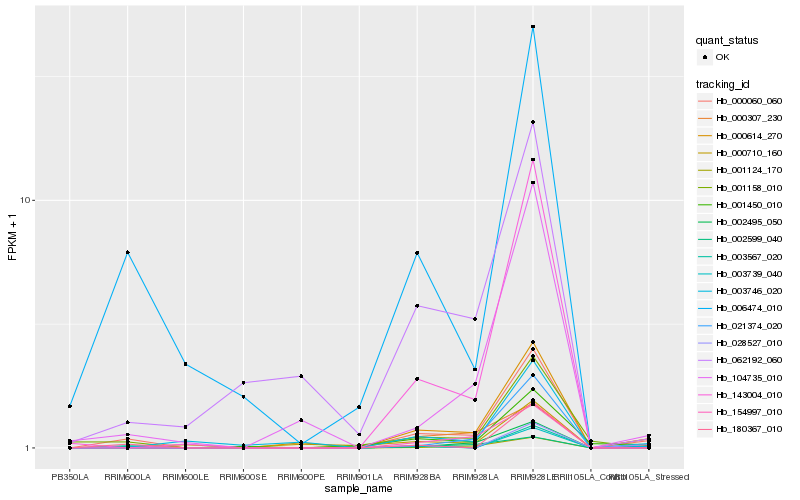
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000060_060 |
0.0 |
transcription factor |
TF Family: C2H2 |
hypothetical protein JCGZ_22940 [Jatropha curcas] |
| 2 |
Hb_000614_270 |
0.1854893622 |
- |
- |
- |
| 3 |
Hb_002599_040 |
0.1974836623 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 4 |
Hb_104735_010 |
0.2151745835 |
- |
- |
cytochrome c biogenesis C (mitochondrion) [Hevea brasiliensis] |
| 5 |
Hb_143004_010 |
0.2223419468 |
- |
- |
PREDICTED: uncharacterized J domain-containing protein C1071.09c [Eucalyptus grandis] |
| 6 |
Hb_003739_040 |
0.2300238352 |
- |
- |
RNA-directed DNA polymerase (Reverse transcriptase); Chromo; Zinc finger, CCHC-type; Peptidase aspartic, active site; Polynucleotidyl transferase, Ribonuclease H fold [Medicago truncatula] |
| 7 |
Hb_006474_010 |
0.231303526 |
- |
- |
hypothetical protein VITISV_032357 [Vitis vinifera] |
| 8 |
Hb_003567_020 |
0.236216658 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_180367_010 |
0.2381710046 |
- |
- |
PREDICTED: uncharacterized protein LOC105794100 [Gossypium raimondii] |
| 10 |
Hb_021374_020 |
0.2401053772 |
- |
- |
PREDICTED: uncharacterized protein LOC101298847, partial [Fragaria vesca subsp. vesca] |
| 11 |
Hb_154997_010 |
0.2423583409 |
- |
- |
hypothetical protein POPTR_0001s43680g, partial [Populus trichocarpa] |
| 12 |
Hb_002495_050 |
0.2440443608 |
- |
- |
PREDICTED: uncharacterized protein LOC105639161 [Jatropha curcas] |
| 13 |
Hb_001124_170 |
0.2448057995 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |
| 14 |
Hb_001450_010 |
0.2477567958 |
- |
- |
PREDICTED: uncharacterized protein LOC105436302 [Cucumis sativus] |
| 15 |
Hb_001158_010 |
0.2491061691 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 16 |
Hb_003746_020 |
0.2518655644 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlH, chloroplastic [Gossypium raimondii] |
| 17 |
Hb_062192_060 |
0.2527426296 |
- |
- |
maturase (mitochondrion) [Hevea brasiliensis] |
| 18 |
Hb_000710_160 |
0.2563281802 |
- |
- |
hypothetical protein POPTR_0011s01952g [Populus trichocarpa] |
| 19 |
Hb_028527_010 |
0.2569458271 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 20 |
Hb_000307_230 |
0.26373794 |
- |
- |
- |