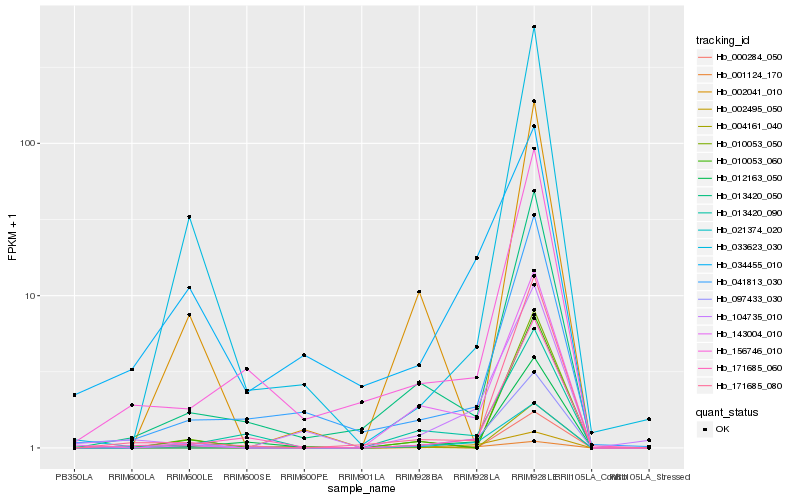
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_097433_030 |
0.0 |
- |
- |
hypothetical protein CISIN_1g0017192mg, partial [Citrus sinensis] |
| 2 |
Hb_021374_020 |
0.1302668854 |
- |
- |
PREDICTED: uncharacterized protein LOC101298847, partial [Fragaria vesca subsp. vesca] |
| 3 |
Hb_143004_010 |
0.1579696 |
- |
- |
PREDICTED: uncharacterized J domain-containing protein C1071.09c [Eucalyptus grandis] |
| 4 |
Hb_033623_030 |
0.1718093394 |
- |
- |
PREDICTED: isoprene synthase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_013420_050 |
0.1734888067 |
- |
- |
hypothetical protein (mitochondrion) [Vicia faba] |
| 6 |
Hb_171685_080 |
0.173609243 |
- |
- |
unknown [Allium cepa] |
| 7 |
Hb_012163_050 |
0.1765472487 |
- |
- |
ATPase subunit [Medicago truncatula] |
| 8 |
Hb_001124_170 |
0.1785146219 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |
| 9 |
Hb_002495_050 |
0.1793023517 |
- |
- |
PREDICTED: uncharacterized protein LOC105639161 [Jatropha curcas] |
| 10 |
Hb_000284_050 |
0.1807588453 |
- |
- |
hypothetical protein L484_026849 [Morus notabilis] |
| 11 |
Hb_004161_040 |
0.1817663044 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Populus euphratica] |
| 12 |
Hb_171685_060 |
0.1817854819 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 2 [Pyrus x bretschneideri] |
| 13 |
Hb_104735_010 |
0.1834788357 |
- |
- |
cytochrome c biogenesis C (mitochondrion) [Hevea brasiliensis] |
| 14 |
Hb_041813_030 |
0.1836773676 |
- |
- |
cytochrome oxidase subunit 1, partial (mitochondrion) [Calochone redingii] |
| 15 |
Hb_010053_050 |
0.184163363 |
- |
- |
hypothetical protein POPTR_0530s002102g, partial [Populus trichocarpa] |
| 16 |
Hb_010053_060 |
0.1882064352 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 17 |
Hb_013420_090 |
0.1903300892 |
- |
- |
BnaCnng13090D [Brassica napus] |
| 18 |
Hb_034455_010 |
0.1953059497 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 19 |
Hb_002041_010 |
0.1958158728 |
- |
- |
- |
| 20 |
Hb_156746_010 |
0.1959229245 |
- |
- |
ATP synthase F1 subunit 1 (mitochondrion) [Hevea brasiliensis] |