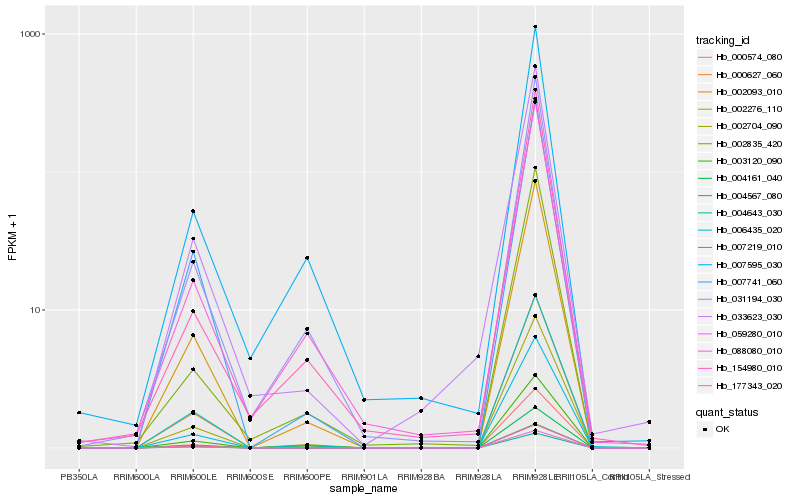
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_033623_030 |
0.0 |
- |
- |
PREDICTED: isoprene synthase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 2 |
Hb_000627_060 |
0.0772265467 |
- |
- |
- |
| 3 |
Hb_002704_090 |
0.0788282674 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 4 |
Hb_002093_010 |
0.0792310999 |
- |
- |
PREDICTED: non-symbiotic hemoglobin 2 [Jatropha curcas] |
| 5 |
Hb_006435_020 |
0.0794165578 |
- |
- |
sugar transporter, putative [Ricinus communis] |
| 6 |
Hb_031194_030 |
0.0809187969 |
- |
- |
ribulose 1,5-bisphosphate carboxylase/oxygenase [Stackhousia minima] |
| 7 |
Hb_007741_060 |
0.0810926656 |
- |
- |
PREDICTED: putative lipid-binding protein At4g00165 [Jatropha curcas] |
| 8 |
Hb_002276_110 |
0.0825914348 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 9 |
Hb_059280_010 |
0.0827685574 |
- |
- |
hypothetical protein POPTR_0019s02860g [Populus trichocarpa] |
| 10 |
Hb_088080_010 |
0.0828761986 |
- |
- |
photosystem I P700 chlorophyll a apoprotein A1 [Matelea biflora] |
| 11 |
Hb_007219_010 |
0.0836258276 |
- |
- |
- |
| 12 |
Hb_004567_080 |
0.0871396342 |
- |
- |
Xyloglucan endotransglucosylase/hydrolase protein 2 precursor, putative [Ricinus communis] |
| 13 |
Hb_004643_030 |
0.0871596204 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_002835_420 |
0.0877620761 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 15 |
Hb_177343_020 |
0.0894218841 |
- |
- |
PREDICTED: uncharacterized protein LOC104751876 [Camelina sativa] |
| 16 |
Hb_007595_030 |
0.0897394173 |
- |
- |
psbA gene product (chloroplast) [Silene noctiflora] |
| 17 |
Hb_004161_040 |
0.0904542037 |
- |
- |
PREDICTED: putative receptor-like protein kinase At4g00960 [Populus euphratica] |
| 18 |
Hb_154980_010 |
0.0916557311 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 19 |
Hb_003120_090 |
0.0939179522 |
- |
- |
PREDICTED: uncharacterized protein LOC100264374 [Vitis vinifera] |
| 20 |
Hb_000574_080 |
0.0939610828 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |