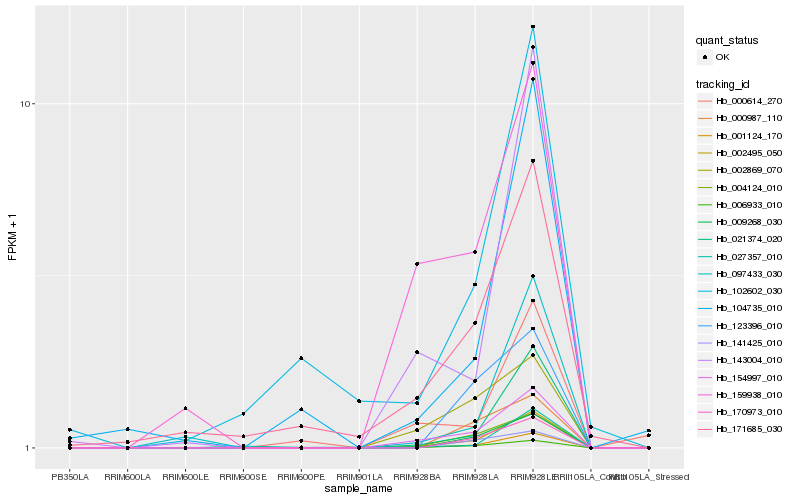
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002495_050 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105639161 [Jatropha curcas] |
| 2 |
Hb_001124_170 |
0.0024726215 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |
| 3 |
Hb_006933_010 |
0.088035048 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_154997_010 |
0.0939539903 |
- |
- |
hypothetical protein POPTR_0001s43680g, partial [Populus trichocarpa] |
| 5 |
Hb_021374_020 |
0.098631893 |
- |
- |
PREDICTED: uncharacterized protein LOC101298847, partial [Fragaria vesca subsp. vesca] |
| 6 |
Hb_004124_010 |
0.1164174149 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 7 |
Hb_170973_010 |
0.1516282529 |
- |
- |
PREDICTED: uncharacterized protein LOC105644346 [Jatropha curcas] |
| 8 |
Hb_009268_030 |
0.1516919008 |
- |
- |
ribosomal protein S7 [Piper cenocladum] |
| 9 |
Hb_097433_030 |
0.1793023517 |
- |
- |
hypothetical protein CISIN_1g0017192mg, partial [Citrus sinensis] |
| 10 |
Hb_143004_010 |
0.1799138147 |
- |
- |
PREDICTED: uncharacterized J domain-containing protein C1071.09c [Eucalyptus grandis] |
| 11 |
Hb_002869_070 |
0.1928683073 |
- |
- |
- |
| 12 |
Hb_159938_010 |
0.196880622 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 13 |
Hb_171685_030 |
0.1972671021 |
- |
- |
URF-1 [Citrullus lanatus] |
| 14 |
Hb_104735_010 |
0.201533014 |
- |
- |
cytochrome c biogenesis C (mitochondrion) [Hevea brasiliensis] |
| 15 |
Hb_141425_010 |
0.202778702 |
- |
- |
hypothetical protein POPTR_0001s43680g, partial [Populus trichocarpa] |
| 16 |
Hb_000987_110 |
0.2042133613 |
- |
- |
PREDICTED: uncharacterized protein LOC104902780 [Beta vulgaris subsp. vulgaris] |
| 17 |
Hb_123396_010 |
0.208013273 |
- |
- |
hypothetical protein AMTR_s00510p00011440 [Amborella trichopoda] |
| 18 |
Hb_027357_010 |
0.2226201773 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_102602_030 |
0.223023702 |
- |
- |
hypothetical protein (mitochondrion) [Vicia faba] |
| 20 |
Hb_000614_270 |
0.2261122485 |
- |
- |
- |