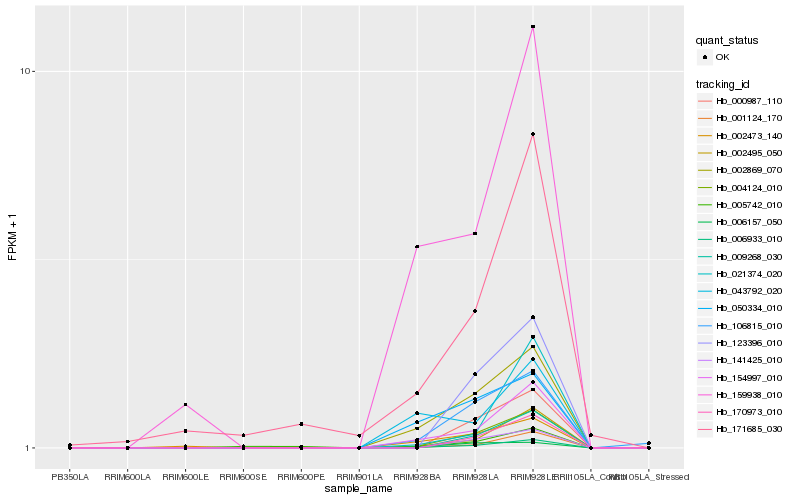
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006933_010 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_004124_010 |
0.0526476225 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 3 |
Hb_154997_010 |
0.0737813157 |
- |
- |
hypothetical protein POPTR_0001s43680g, partial [Populus trichocarpa] |
| 4 |
Hb_002495_050 |
0.088035048 |
- |
- |
PREDICTED: uncharacterized protein LOC105639161 [Jatropha curcas] |
| 5 |
Hb_001124_170 |
0.0892301693 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |
| 6 |
Hb_002869_070 |
0.1109103441 |
- |
- |
- |
| 7 |
Hb_170973_010 |
0.1516976833 |
- |
- |
PREDICTED: uncharacterized protein LOC105644346 [Jatropha curcas] |
| 8 |
Hb_009268_030 |
0.1516982073 |
- |
- |
ribosomal protein S7 [Piper cenocladum] |
| 9 |
Hb_106815_010 |
0.1631054838 |
- |
- |
hypothetical protein, conserved [Eimeria acervulina] |
| 10 |
Hb_141425_010 |
0.1695353423 |
- |
- |
hypothetical protein POPTR_0001s43680g, partial [Populus trichocarpa] |
| 11 |
Hb_000987_110 |
0.1703667553 |
- |
- |
PREDICTED: uncharacterized protein LOC104902780 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_123396_010 |
0.1726271577 |
- |
- |
hypothetical protein AMTR_s00510p00011440 [Amborella trichopoda] |
| 13 |
Hb_159938_010 |
0.1761048897 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |
| 14 |
Hb_002473_140 |
0.1827322956 |
- |
- |
- |
| 15 |
Hb_021374_020 |
0.1852993264 |
- |
- |
PREDICTED: uncharacterized protein LOC101298847, partial [Fragaria vesca subsp. vesca] |
| 16 |
Hb_171685_030 |
0.1953694763 |
- |
- |
URF-1 [Citrullus lanatus] |
| 17 |
Hb_050334_010 |
0.2051014965 |
- |
- |
- |
| 18 |
Hb_005742_010 |
0.2212255851 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 19 |
Hb_043792_020 |
0.2225369691 |
- |
- |
Retrotransposon gag protein [Medicago truncatula] |
| 20 |
Hb_006157_050 |
0.2229742352 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |