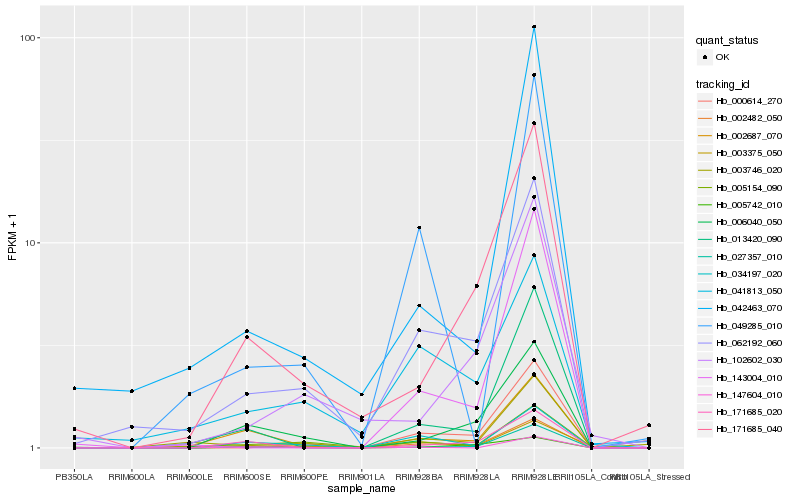
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002482_050 |
0.0 |
transcription factor |
TF Family: NAC |
F-box family protein [Populus trichocarpa] |
| 2 |
Hb_062192_060 |
0.1269978444 |
- |
- |
maturase (mitochondrion) [Hevea brasiliensis] |
| 3 |
Hb_034197_020 |
0.1600259973 |
- |
- |
hypothetical protein VITISV_041099 [Vitis vinifera] |
| 4 |
Hb_013420_090 |
0.1676287182 |
- |
- |
BnaCnng13090D [Brassica napus] |
| 5 |
Hb_006040_050 |
0.1738486665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_027357_010 |
0.1751855896 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_002687_070 |
0.1794622452 |
- |
- |
polyprotein [Ananas comosus] |
| 8 |
Hb_003375_050 |
0.1842378828 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 9 |
Hb_171685_040 |
0.1869487037 |
- |
- |
ribosomal protein S13 (mitochondrion) [Hevea brasiliensis] |
| 10 |
Hb_000614_270 |
0.1949559139 |
- |
- |
- |
| 11 |
Hb_171685_020 |
0.1960583626 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_005742_010 |
0.1982313521 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 13 |
Hb_005154_090 |
0.1991857678 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_003746_020 |
0.2042263979 |
- |
- |
PREDICTED: magnesium-chelatase subunit ChlH, chloroplastic [Gossypium raimondii] |
| 15 |
Hb_143004_010 |
0.2056983647 |
- |
- |
PREDICTED: uncharacterized J domain-containing protein C1071.09c [Eucalyptus grandis] |
| 16 |
Hb_049285_010 |
0.2084705811 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 17 |
Hb_102602_030 |
0.2131399289 |
- |
- |
hypothetical protein (mitochondrion) [Vicia faba] |
| 18 |
Hb_042463_070 |
0.2135688307 |
- |
- |
60S ribosomal protein L16, putative [Ricinus communis] |
| 19 |
Hb_041813_050 |
0.2143322749 |
- |
- |
hypothetical protein NitaMp096 [Nicotiana tabacum] |
| 20 |
Hb_147604_010 |
0.2178274423 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |