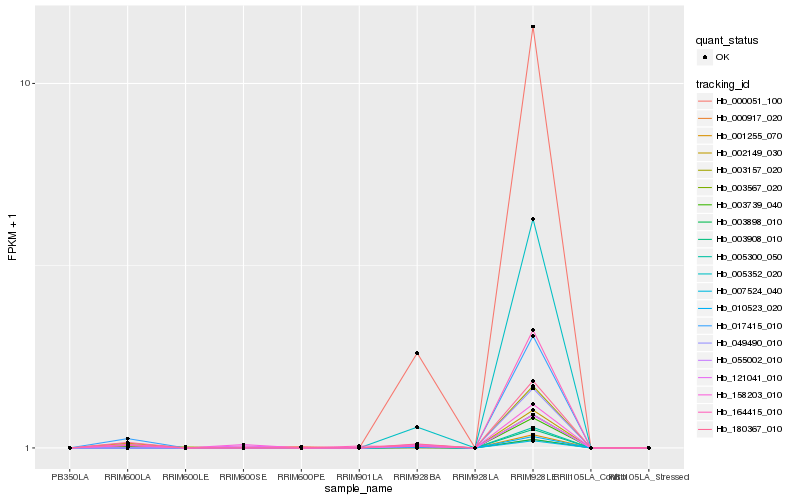
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_180367_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105794100 [Gossypium raimondii] |
| 2 |
Hb_003567_020 |
0.0406665972 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_003739_040 |
0.081760046 |
- |
- |
RNA-directed DNA polymerase (Reverse transcriptase); Chromo; Zinc finger, CCHC-type; Peptidase aspartic, active site; Polynucleotidyl transferase, Ribonuclease H fold [Medicago truncatula] |
| 4 |
Hb_017415_010 |
0.0899099322 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 5 |
Hb_003157_020 |
0.1448462197 |
- |
- |
PREDICTED: uncharacterized protein LOC104809111 [Tarenaya hassleriana] |
| 6 |
Hb_002149_030 |
0.1601131406 |
- |
- |
PREDICTED: uncharacterized protein LOC105647984 [Jatropha curcas] |
| 7 |
Hb_164415_010 |
0.1620465435 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_121041_010 |
0.1667515977 |
- |
- |
PREDICTED: uncharacterized protein LOC105778857 [Gossypium raimondii] |
| 9 |
Hb_000051_100 |
0.1704297209 |
- |
- |
hypothetical protein MIMGU_mgv1a025286mg [Erythranthe guttata] |
| 10 |
Hb_158203_010 |
0.1711337038 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 11 |
Hb_005300_050 |
0.1755468696 |
- |
- |
hypothetical protein VITISV_001308 [Vitis vinifera] |
| 12 |
Hb_003908_010 |
0.1755514923 |
- |
- |
polyprotein [Oryza australiensis] |
| 13 |
Hb_001255_070 |
0.1755660217 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 14 |
Hb_000917_020 |
0.1755810953 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 15 |
Hb_007524_040 |
0.1755855089 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC101511096 [Cicer arietinum] |
| 16 |
Hb_005352_020 |
0.1757894089 |
- |
- |
- |
| 17 |
Hb_049490_010 |
0.1763093439 |
- |
- |
hypothetical protein VITISV_013805 [Vitis vinifera] |
| 18 |
Hb_055002_010 |
0.1766727874 |
- |
- |
PREDICTED: uncharacterized protein LOC104907627 [Beta vulgaris subsp. vulgaris] |
| 19 |
Hb_003898_010 |
0.1769467031 |
- |
- |
hypothetical protein PRUPE_ppa022673mg [Prunus persica] |
| 20 |
Hb_010523_020 |
0.1770226661 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |