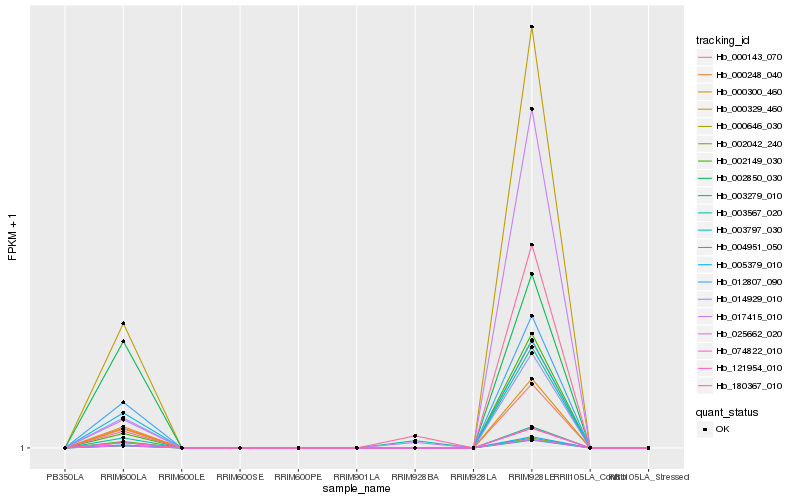
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000329_460 |
0.0 |
- |
- |
PREDICTED: protein transport protein Sec61 subunit beta [Cucumis sativus] |
| 2 |
Hb_000248_040 |
0.0237050667 |
- |
- |
PREDICTED: uncharacterized protein LOC105632434 [Jatropha curcas] |
| 3 |
Hb_003797_030 |
0.0360194266 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_025662_020 |
0.0361560284 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_000646_030 |
0.0361891667 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 6 |
Hb_005379_010 |
0.0367204067 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 7 |
Hb_074822_010 |
0.0384889059 |
- |
- |
PREDICTED: uncharacterized protein LOC104088736 isoform X2 [Nicotiana tomentosiformis] |
| 8 |
Hb_121954_010 |
0.0385661329 |
- |
- |
PREDICTED: uncharacterized protein LOC105774125 [Gossypium raimondii] |
| 9 |
Hb_003279_010 |
0.0388362817 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 10 |
Hb_000300_460 |
0.0420564663 |
- |
- |
Mavicyanin, putative [Ricinus communis] |
| 11 |
Hb_000143_070 |
0.0487519633 |
- |
- |
- |
| 12 |
Hb_014929_010 |
0.0569901311 |
- |
- |
- |
| 13 |
Hb_004951_050 |
0.0607585978 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_012807_090 |
0.0684847444 |
- |
- |
PREDICTED: uncharacterized protein LOC100267173 [Vitis vinifera] |
| 15 |
Hb_002149_030 |
0.0887954502 |
- |
- |
PREDICTED: uncharacterized protein LOC105647984 [Jatropha curcas] |
| 16 |
Hb_017415_010 |
0.176344576 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 17 |
Hb_002850_030 |
0.1823743082 |
- |
- |
- |
| 18 |
Hb_003567_020 |
0.2051028913 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_180367_010 |
0.2085658002 |
- |
- |
PREDICTED: uncharacterized protein LOC105794100 [Gossypium raimondii] |
| 20 |
Hb_002042_240 |
0.2086625982 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |