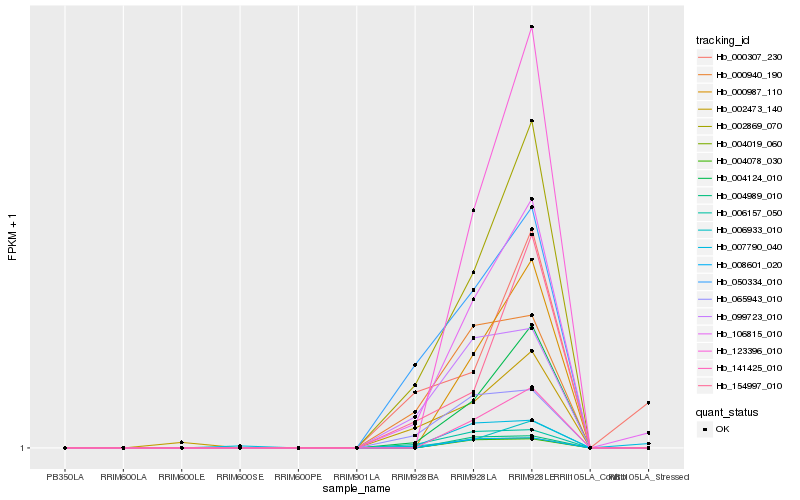
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_106815_010 |
0.0 |
- |
- |
hypothetical protein, conserved [Eimeria acervulina] |
| 2 |
Hb_002869_070 |
0.1445711093 |
- |
- |
- |
| 3 |
Hb_004124_010 |
0.1504108055 |
- |
- |
PREDICTED: uncharacterized protein LOC105631113 [Jatropha curcas] |
| 4 |
Hb_006933_010 |
0.1631054838 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 5 |
Hb_006157_050 |
0.1722357632 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_065943_010 |
0.1777726884 |
- |
- |
- |
| 7 |
Hb_099723_010 |
0.1878858743 |
- |
- |
hypothetical protein JCGZ_15340 [Jatropha curcas] |
| 8 |
Hb_007790_040 |
0.1879361189 |
- |
- |
PREDICTED: uncharacterized protein LOC104417177 [Eucalyptus grandis] |
| 9 |
Hb_000940_190 |
0.1906851836 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 10 |
Hb_050334_010 |
0.1967693343 |
- |
- |
- |
| 11 |
Hb_123396_010 |
0.1987158454 |
- |
- |
hypothetical protein AMTR_s00510p00011440 [Amborella trichopoda] |
| 12 |
Hb_000987_110 |
0.1994775138 |
- |
- |
PREDICTED: uncharacterized protein LOC104902780 [Beta vulgaris subsp. vulgaris] |
| 13 |
Hb_141425_010 |
0.1997970169 |
- |
- |
hypothetical protein POPTR_0001s43680g, partial [Populus trichocarpa] |
| 14 |
Hb_154997_010 |
0.20279669 |
- |
- |
hypothetical protein POPTR_0001s43680g, partial [Populus trichocarpa] |
| 15 |
Hb_002473_140 |
0.2058337023 |
- |
- |
- |
| 16 |
Hb_000307_230 |
0.2139930989 |
- |
- |
- |
| 17 |
Hb_004078_030 |
0.2222224647 |
- |
- |
Putative polyprotein, identical [Solanum demissum] |
| 18 |
Hb_004019_060 |
0.2222322415 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 19 |
Hb_008601_020 |
0.2222350304 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 20 |
Hb_004989_010 |
0.2222583138 |
- |
- |
retrotransposon protein, putative, Ty3-gypsy sub-class [Oryza sativa Japonica Group] |