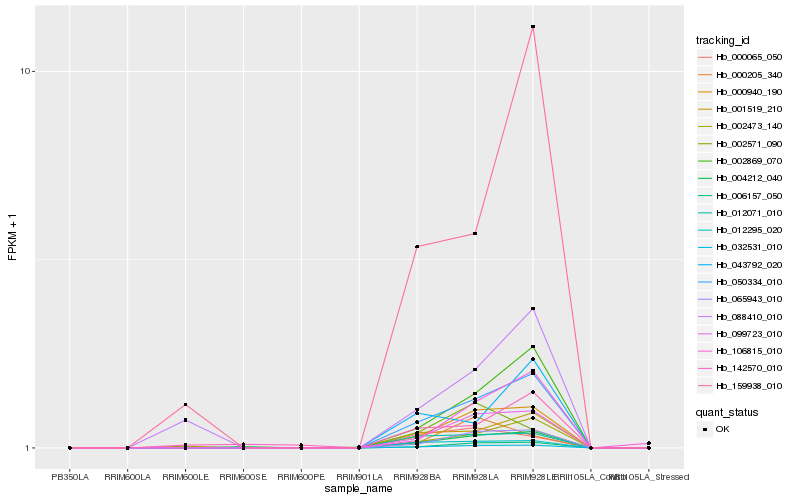
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032531_010 |
0.0 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 2 |
Hb_000940_190 |
0.0597885837 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 3 |
Hb_099723_010 |
0.0647757189 |
- |
- |
hypothetical protein JCGZ_15340 [Jatropha curcas] |
| 4 |
Hb_050334_010 |
0.0762775691 |
- |
- |
- |
| 5 |
Hb_065943_010 |
0.0840355844 |
- |
- |
- |
| 6 |
Hb_006157_050 |
0.0956830686 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 7 |
Hb_012295_020 |
0.1255808245 |
- |
- |
PREDICTED: uncharacterized protein LOC104223093 [Nicotiana sylvestris] |
| 8 |
Hb_012071_010 |
0.13450572 |
- |
- |
PREDICTED: uncharacterized protein LOC105629655 [Jatropha curcas] |
| 9 |
Hb_001519_210 |
0.1409724054 |
- |
- |
hypothetical protein B456_003G032500 [Gossypium raimondii] |
| 10 |
Hb_002869_070 |
0.1576867753 |
- |
- |
- |
| 11 |
Hb_004212_040 |
0.1662977045 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 12 |
Hb_000065_050 |
0.1848653916 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 13 |
Hb_002473_140 |
0.1896157313 |
- |
- |
- |
| 14 |
Hb_002571_090 |
0.2055649286 |
- |
- |
- |
| 15 |
Hb_106815_010 |
0.2253099208 |
- |
- |
hypothetical protein, conserved [Eimeria acervulina] |
| 16 |
Hb_000205_340 |
0.22619264 |
- |
- |
PREDICTED: putative B3 domain-containing protein At2g27410 [Jatropha curcas] |
| 17 |
Hb_088410_010 |
0.2412435356 |
- |
- |
PREDICTED: choline monooxygenase, chloroplastic isoform X2 [Jatropha curcas] |
| 18 |
Hb_043792_020 |
0.243150302 |
- |
- |
Retrotransposon gag protein [Medicago truncatula] |
| 19 |
Hb_142570_010 |
0.2593146411 |
- |
- |
PREDICTED: uncharacterized protein LOC104907573 [Beta vulgaris subsp. vulgaris] |
| 20 |
Hb_159938_010 |
0.2627302436 |
- |
- |
phosphoprotein phosphatase, putative [Ricinus communis] |