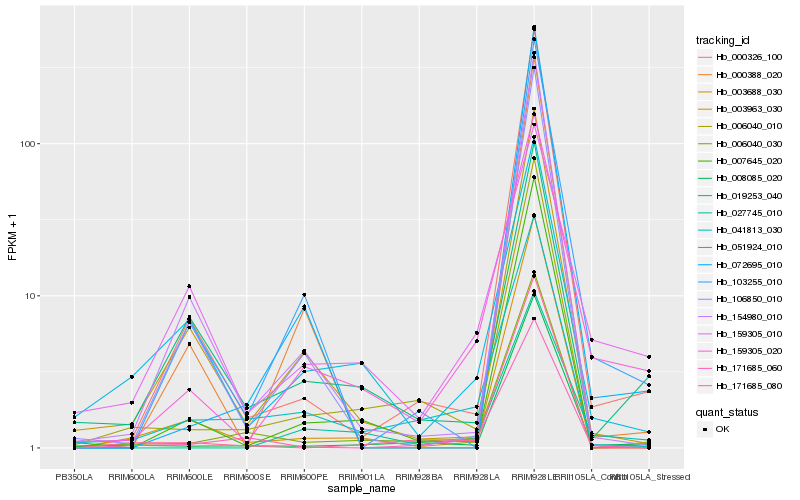
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_159305_020 |
0.0 |
- |
- |
50S ribosomal protein L2-B [Medicago truncatula] |
| 2 |
Hb_027745_010 |
0.106182858 |
- |
- |
50S ribosomal protein L2, putative [Ricinus communis] |
| 3 |
Hb_008085_020 |
0.1318093729 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At5g49770 [Populus euphratica] |
| 4 |
Hb_041813_030 |
0.1477578177 |
- |
- |
cytochrome oxidase subunit 1, partial (mitochondrion) [Calochone redingii] |
| 5 |
Hb_103255_010 |
0.1486680099 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 6 |
Hb_007645_020 |
0.1530406369 |
- |
- |
NAD(P)H-quinone oxidoreductase subunit [Medicago truncatula] |
| 7 |
Hb_006040_010 |
0.1572030061 |
- |
- |
cytochrome c oxidase subunit 3 (mitochondrion) [Hevea brasiliensis] |
| 8 |
Hb_006040_030 |
0.160882404 |
- |
- |
hypothetical protein EUTSA_v10001274mg [Eutrema salsugineum] |
| 9 |
Hb_159305_010 |
0.1610152337 |
- |
- |
rpl2 [Jatropha curcas] |
| 10 |
Hb_000326_100 |
0.1620117228 |
transcription factor |
TF Family: MYB-related |
DnaJ homolog subfamily C member 2 [Morus notabilis] |
| 11 |
Hb_003963_030 |
0.1661421585 |
- |
- |
psbB [Jatropha curcas] |
| 12 |
Hb_003688_030 |
0.1667727402 |
- |
- |
NADH dehydrogenase 2-like ORF 180 |
| 13 |
Hb_051924_010 |
0.1669333454 |
- |
- |
BnaA01g34100D [Brassica napus] |
| 14 |
Hb_171685_080 |
0.1735316851 |
- |
- |
unknown [Allium cepa] |
| 15 |
Hb_072695_010 |
0.173546354 |
- |
- |
PREDICTED: metalloendoproteinase 1-like [Populus euphratica] |
| 16 |
Hb_154980_010 |
0.1741815157 |
- |
- |
photosystem I P700 chlorophyll A apoprotein A2 [Medicago truncatula] |
| 17 |
Hb_019253_040 |
0.1743720818 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_171685_060 |
0.1759546042 |
- |
- |
PREDICTED: cytochrome c oxidase subunit 2 [Pyrus x bretschneideri] |
| 19 |
Hb_106850_010 |
0.176722924 |
- |
- |
photosystem II protein D2 [Lactuca sativa] |
| 20 |
Hb_000388_020 |
0.1771821722 |
- |
- |
PREDICTED: MLP-like protein 43 [Fragaria vesca subsp. vesca] |