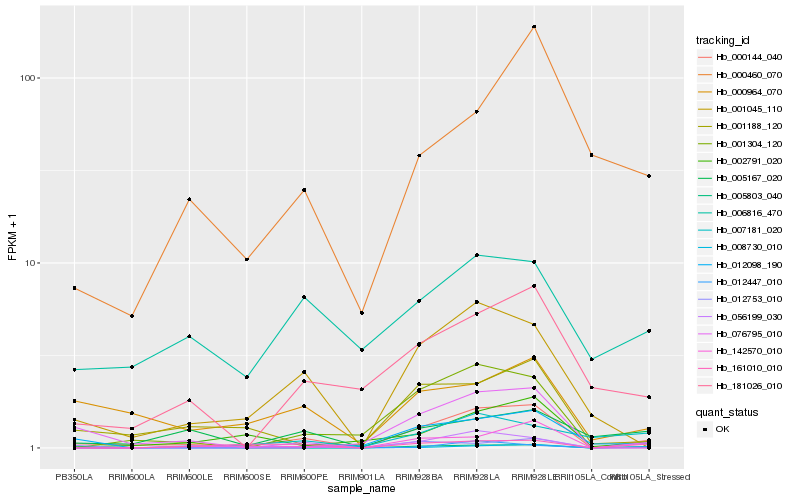
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000144_040 |
0.0 |
- |
- |
hypothetical protein POPTR_0006s06790g [Populus trichocarpa] |
| 2 |
Hb_001304_120 |
0.2362030781 |
transcription factor |
TF Family: WRKY |
WRKY protein [Hevea brasiliensis] |
| 3 |
Hb_001045_110 |
0.2421440496 |
transcription factor |
TF Family: M-type |
flowering locus C-like protein [Juglans regia] |
| 4 |
Hb_001188_120 |
0.2525308417 |
- |
- |
hypothetical protein JCGZ_05575 [Jatropha curcas] |
| 5 |
Hb_076795_010 |
0.2528566891 |
- |
- |
hypothetical protein POPTR_0001s07680g, partial [Populus trichocarpa] |
| 6 |
Hb_012098_190 |
0.2546232974 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance RPP13-like protein 1 [Jatropha curcas] |
| 7 |
Hb_056199_030 |
0.261847255 |
- |
- |
PREDICTED: purine permease 3-like isoform X2 [Gossypium raimondii] |
| 8 |
Hb_181026_010 |
0.2685210797 |
- |
- |
Auxin-induced protein 5NG4, putative [Ricinus communis] |
| 9 |
Hb_000964_070 |
0.2731072347 |
- |
- |
PREDICTED: ATP synthase subunit O, mitochondrial [Jatropha curcas] |
| 10 |
Hb_002791_020 |
0.2754314663 |
- |
- |
hypothetical protein AALP_AA5G271700 [Arabis alpina] |
| 11 |
Hb_142570_010 |
0.2893823269 |
- |
- |
PREDICTED: uncharacterized protein LOC104907573 [Beta vulgaris subsp. vulgaris] |
| 12 |
Hb_005167_020 |
0.2906855034 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 13 |
Hb_000460_070 |
0.2958758326 |
- |
- |
luminal binding protein [Gossypium hirsutum] |
| 14 |
Hb_005803_040 |
0.2984804571 |
- |
- |
PREDICTED: uncharacterized protein LOC104908276 [Beta vulgaris subsp. vulgaris] |
| 15 |
Hb_161010_010 |
0.3005065855 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 16 |
Hb_012447_010 |
0.3014897368 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 17 |
Hb_008730_010 |
0.3028433149 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_012753_010 |
0.3075893775 |
- |
- |
PREDICTED: uncharacterized protein LOC105636491 [Jatropha curcas] |
| 19 |
Hb_007181_020 |
0.3082799698 |
- |
- |
PREDICTED: uncharacterized protein At4g04980-like [Populus euphratica] |
| 20 |
Hb_006816_470 |
0.3083544151 |
- |
- |
PREDICTED: uncharacterized protein LOC105647469 isoform X1 [Jatropha curcas] |