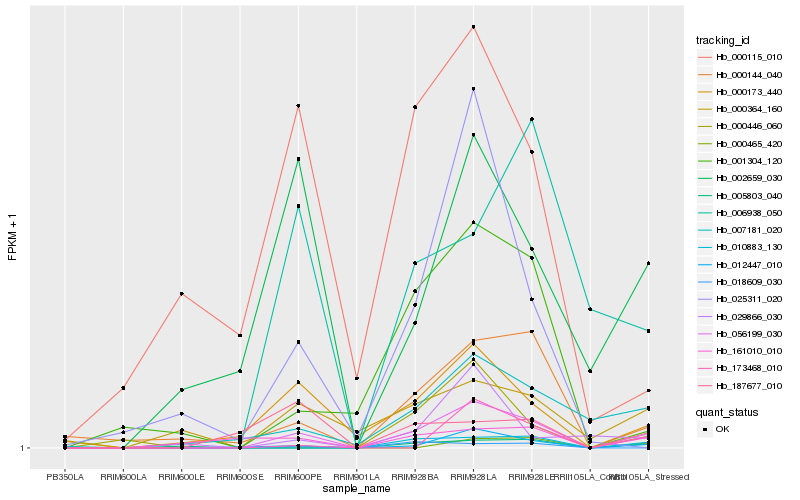
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_056199_030 |
0.0 |
- |
- |
PREDICTED: purine permease 3-like isoform X2 [Gossypium raimondii] |
| 2 |
Hb_012447_010 |
0.2076548892 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 3 |
Hb_000144_040 |
0.261847255 |
- |
- |
hypothetical protein POPTR_0006s06790g [Populus trichocarpa] |
| 4 |
Hb_000173_440 |
0.2635394155 |
- |
- |
YALI0B05280p [Yarrowia lipolytica] |
| 5 |
Hb_007181_020 |
0.2815103821 |
- |
- |
PREDICTED: uncharacterized protein At4g04980-like [Populus euphratica] |
| 6 |
Hb_010883_130 |
0.2934967138 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g08210 [Jatropha curcas] |
| 7 |
Hb_001304_120 |
0.3003363788 |
transcription factor |
TF Family: WRKY |
WRKY protein [Hevea brasiliensis] |
| 8 |
Hb_002659_030 |
0.3039337566 |
- |
- |
copper transport protein atox1, putative [Ricinus communis] |
| 9 |
Hb_000364_160 |
0.3108033533 |
- |
- |
PREDICTED: uncharacterized protein LOC105639754 [Jatropha curcas] |
| 10 |
Hb_000465_420 |
0.3120328642 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105632322 [Jatropha curcas] |
| 11 |
Hb_161010_010 |
0.318568953 |
- |
- |
MULTISPECIES: hypothetical protein, partial [Pseudomonas fluorescens group] |
| 12 |
Hb_006938_050 |
0.3192326036 |
- |
- |
PREDICTED: uncharacterized protein LOC105648204 [Jatropha curcas] |
| 13 |
Hb_025311_020 |
0.3241388416 |
- |
- |
PREDICTED: uncharacterized protein LOC105039527 [Elaeis guineensis] |
| 14 |
Hb_173468_010 |
0.32723422 |
- |
- |
Vacuolar sorting receptor 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_005803_040 |
0.3300417816 |
- |
- |
PREDICTED: uncharacterized protein LOC104908276 [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_018609_030 |
0.3309225734 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000446_060 |
0.334293872 |
- |
- |
- |
| 18 |
Hb_000115_010 |
0.3416558721 |
- |
- |
5-methyltetrahydrofolate:homocysteine methyltransferase, putative [Ricinus communis] |
| 19 |
Hb_187677_010 |
0.344808417 |
- |
- |
- |
| 20 |
Hb_029866_030 |
0.3464721551 |
- |
- |
hypothetical protein CICLE_v10006898mg [Citrus clementina] |