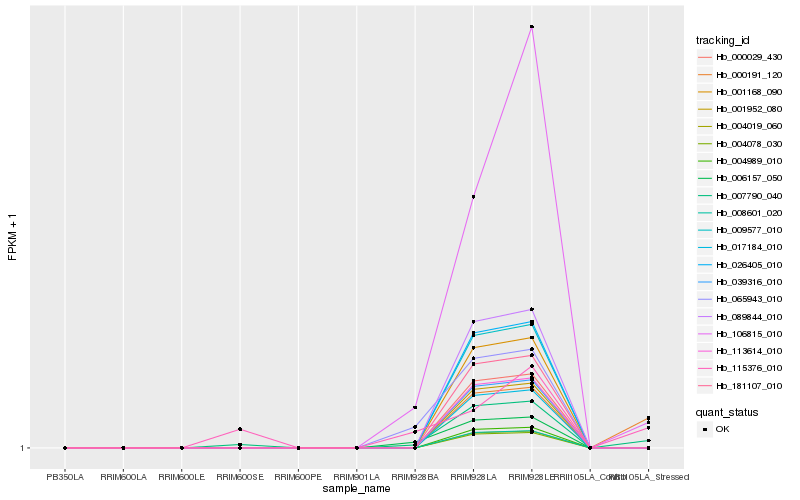
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007790_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104417177 [Eucalyptus grandis] |
| 2 |
Hb_106815_010 |
0.1879361189 |
- |
- |
hypothetical protein, conserved [Eimeria acervulina] |
| 3 |
Hb_115376_010 |
0.2304368506 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 4 |
Hb_000191_120 |
0.2356644252 |
- |
- |
- |
| 5 |
Hb_006157_050 |
0.25350587 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_065943_010 |
0.2581152734 |
- |
- |
- |
| 7 |
Hb_004078_030 |
0.263565254 |
- |
- |
Putative polyprotein, identical [Solanum demissum] |
| 8 |
Hb_004019_060 |
0.2635652542 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 9 |
Hb_008601_020 |
0.2635652546 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 10 |
Hb_004989_010 |
0.2635652628 |
- |
- |
retrotransposon protein, putative, Ty3-gypsy sub-class [Oryza sativa Japonica Group] |
| 11 |
Hb_017184_010 |
0.2635659857 |
- |
- |
PREDICTED: uncharacterized protein LOC105778857 [Gossypium raimondii] |
| 12 |
Hb_001952_080 |
0.2635662445 |
- |
- |
PREDICTED: uncharacterized protein LOC105042236 isoform X2 [Elaeis guineensis] |
| 13 |
Hb_039316_010 |
0.2635663868 |
- |
- |
PREDICTED: uncharacterized protein LOC105771917 [Gossypium raimondii] |
| 14 |
Hb_113614_010 |
0.2635664705 |
- |
- |
hypothetical protein VITISV_001308 [Vitis vinifera] |
| 15 |
Hb_000029_430 |
0.2635666693 |
- |
- |
PREDICTED: adenylate isopentenyltransferase 5, chloroplastic-like [Jatropha curcas] |
| 16 |
Hb_181107_010 |
0.2635677638 |
- |
- |
hypothetical protein, partial [Pseudomonas tolaasii] |
| 17 |
Hb_001168_090 |
0.2635691513 |
- |
- |
PREDICTED: uncharacterized protein LOC104246050 [Nicotiana sylvestris] |
| 18 |
Hb_009577_010 |
0.263570409 |
- |
- |
- |
| 19 |
Hb_026405_010 |
0.2635706882 |
- |
- |
PREDICTED: uncharacterized protein LOC104107896 [Nicotiana tomentosiformis] |
| 20 |
Hb_089844_010 |
0.2635720612 |
- |
- |
- |