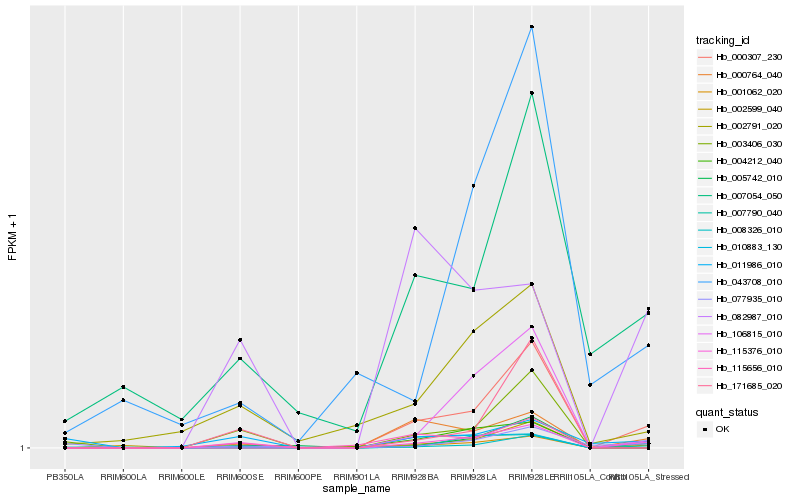
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_115376_010 |
0.0 |
- |
- |
hypothetical protein PRUPE_ppa021229mg [Prunus persica] |
| 2 |
Hb_001062_020 |
0.1724214459 |
- |
- |
PREDICTED: uncharacterized protein LOC103962405 [Pyrus x bretschneideri] |
| 3 |
Hb_007790_040 |
0.2304368506 |
- |
- |
PREDICTED: uncharacterized protein LOC104417177 [Eucalyptus grandis] |
| 4 |
Hb_008326_010 |
0.2457076479 |
- |
- |
putative polyprotein [Oryza sativa Japonica Group] |
| 5 |
Hb_000307_230 |
0.2460480065 |
- |
- |
- |
| 6 |
Hb_002791_020 |
0.2835261358 |
- |
- |
hypothetical protein AALP_AA5G271700 [Arabis alpina] |
| 7 |
Hb_002599_040 |
0.2935660213 |
- |
- |
PREDICTED: uncharacterized protein LOC104105697 [Nicotiana tomentosiformis] |
| 8 |
Hb_003406_030 |
0.3001170501 |
- |
- |
PREDICTED: uncharacterized protein LOC104774100 [Camelina sativa] |
| 9 |
Hb_082987_010 |
0.3005936484 |
- |
- |
hypothetical protein CICLE_v10030423mg [Citrus clementina] |
| 10 |
Hb_106815_010 |
0.3008927524 |
- |
- |
hypothetical protein, conserved [Eimeria acervulina] |
| 11 |
Hb_004212_040 |
0.3091073552 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 12 |
Hb_010883_130 |
0.3099165898 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g08210 [Jatropha curcas] |
| 13 |
Hb_007054_050 |
0.3167036699 |
- |
- |
- |
| 14 |
Hb_171685_020 |
0.3200439271 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000764_040 |
0.3219180156 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 16 |
Hb_043708_010 |
0.322355463 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 17 |
Hb_011986_010 |
0.3244124854 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 18 |
Hb_005742_010 |
0.3246963585 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 19 |
Hb_115656_010 |
0.3251608184 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein RPM1-like [Jatropha curcas] |
| 20 |
Hb_077935_010 |
0.3284764472 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |