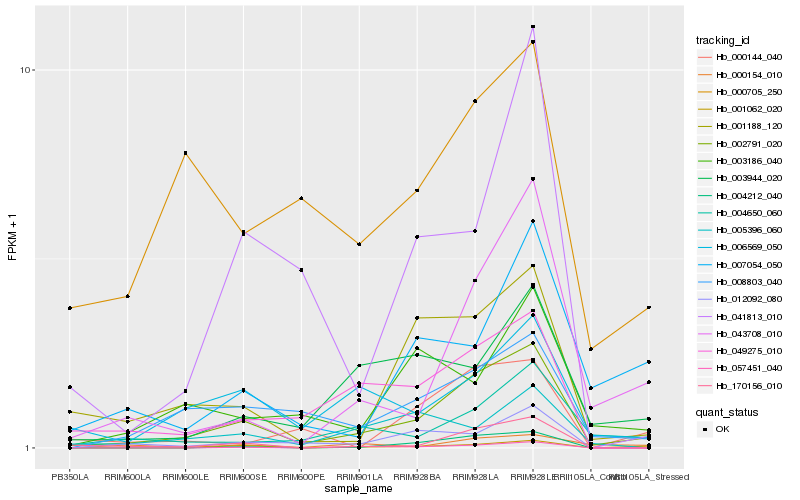
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002791_020 |
0.0 |
- |
- |
hypothetical protein AALP_AA5G271700 [Arabis alpina] |
| 2 |
Hb_008803_040 |
0.2165500377 |
- |
- |
PREDICTED: uncharacterized protein LOC105157106 [Sesamum indicum] |
| 3 |
Hb_043708_010 |
0.2216077134 |
- |
- |
PREDICTED: uncharacterized protein LOC104878039 [Vitis vinifera] |
| 4 |
Hb_170156_010 |
0.232061394 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC104235718, partial [Nicotiana sylvestris] |
| 5 |
Hb_001188_120 |
0.2332456839 |
- |
- |
hypothetical protein JCGZ_05575 [Jatropha curcas] |
| 6 |
Hb_006569_050 |
0.2339050862 |
- |
- |
- |
| 7 |
Hb_004650_060 |
0.2352106373 |
- |
- |
putative retrotransposon protein [Phyllostachys edulis] |
| 8 |
Hb_001062_020 |
0.2454842938 |
- |
- |
PREDICTED: uncharacterized protein LOC103962405 [Pyrus x bretschneideri] |
| 9 |
Hb_049275_010 |
0.2478025122 |
- |
- |
PREDICTED: F-box/LRR-repeat protein 4-like [Populus euphratica] |
| 10 |
Hb_003944_020 |
0.2485606126 |
- |
- |
PREDICTED: uncharacterized protein LOC105110580 [Populus euphratica] |
| 11 |
Hb_012092_080 |
0.2615479876 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g15280-like [Malus domestica] |
| 12 |
Hb_003186_040 |
0.2621075881 |
- |
- |
PREDICTED: protein DENND6A [Jatropha curcas] |
| 13 |
Hb_057451_040 |
0.2649154701 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_004212_040 |
0.2682723605 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 15 |
Hb_007054_050 |
0.2699598895 |
- |
- |
- |
| 16 |
Hb_041813_010 |
0.2702643139 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000144_040 |
0.2754314663 |
- |
- |
hypothetical protein POPTR_0006s06790g [Populus trichocarpa] |
| 18 |
Hb_000154_010 |
0.2765651073 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 19 |
Hb_000705_250 |
0.2785819551 |
- |
- |
hypothetical protein CICLE_v10017607mg [Citrus clementina] |
| 20 |
Hb_005396_060 |
0.2786892464 |
- |
- |
- |