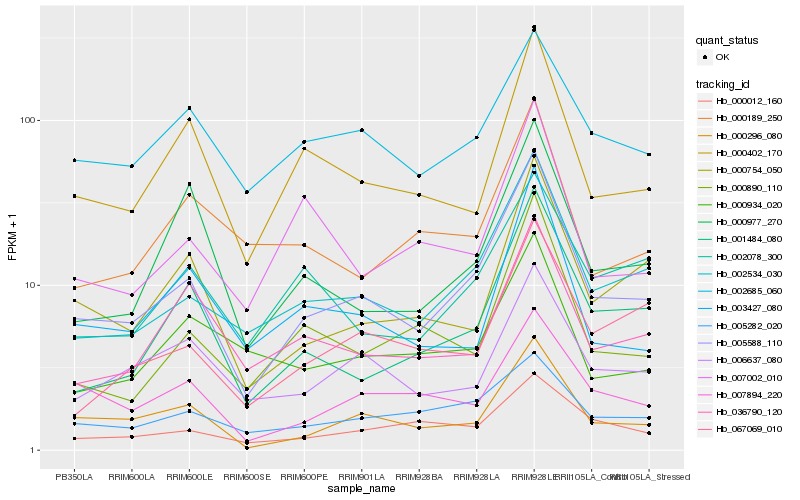
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005588_110 |
0.0 |
- |
- |
hypothetical protein POPTR_0002s12860g [Populus trichocarpa] |
| 2 |
Hb_002534_030 |
0.1010856329 |
transcription factor |
TF Family: M-type |
mads box protein, putative [Ricinus communis] |
| 3 |
Hb_000296_080 |
0.1166828477 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At2g35130 [Jatropha curcas] |
| 4 |
Hb_001484_080 |
0.1431121865 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 5 |
Hb_000754_050 |
0.1455714176 |
- |
- |
PREDICTED: thylakoid lumenal 19 kDa protein, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000402_170 |
0.1469463877 |
- |
- |
PREDICTED: glutamate--glyoxylate aminotransferase 2 [Jatropha curcas] |
| 7 |
Hb_006637_080 |
0.1555180578 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 8 |
Hb_003427_080 |
0.1556419622 |
- |
- |
PREDICTED: probable alanine--tRNA ligase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000890_110 |
0.157505314 |
- |
- |
PREDICTED: GTP-binding protein OBGC, chloroplastic [Populus euphratica] |
| 10 |
Hb_067069_010 |
0.1618181798 |
- |
- |
hypothetical protein JCGZ_11524 [Jatropha curcas] |
| 11 |
Hb_000977_270 |
0.1632506527 |
- |
- |
PREDICTED: phosphoglycolate phosphatase 1B, chloroplastic [Jatropha curcas] |
| 12 |
Hb_007894_220 |
0.1632587508 |
- |
- |
PREDICTED: probable folate-biopterin transporter 4 [Jatropha curcas] |
| 13 |
Hb_002685_060 |
0.1635321407 |
- |
- |
30S ribosomal protein S5, putative [Ricinus communis] |
| 14 |
Hb_036790_120 |
0.1638635166 |
- |
- |
PREDICTED: uncharacterized protein LOC105645122 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000012_160 |
0.1641863623 |
- |
- |
PREDICTED: uncharacterized protein LOC105638175 isoform X1 [Jatropha curcas] |
| 16 |
Hb_005282_020 |
0.1659237655 |
- |
- |
hypothetical protein PRUPE_ppa019056mg, partial [Prunus persica] |
| 17 |
Hb_002078_300 |
0.1710783225 |
- |
- |
PREDICTED: uncharacterized protein LOC105644817 isoform X1 [Jatropha curcas] |
| 18 |
Hb_007002_010 |
0.1711710747 |
- |
- |
PREDICTED: dnaJ protein ERDJ3B [Jatropha curcas] |
| 19 |
Hb_000189_250 |
0.171350098 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000934_020 |
0.1730425775 |
- |
- |
PREDICTED: uncharacterized protein LOC105629153 [Jatropha curcas] |